Date: Thu, 5 Oct 2017 14:56:48 +0530
Hi,
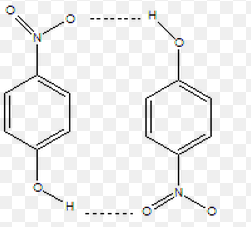
I am trying to mimic inter H-bonds formed in p-nitrophenol dimer.
I drew two p-nitrophenol molecules and placed them 4Å away from each other
in a chloroform box (I have tried many solvent systems) and my expected
output is a structure showing two inter h-bonds between these two molecules
(attached image). However, after running 100-200ns simulations by keeping
them 4Å away ( I have scanned 4Å- 7Å range in different sets) and also in
different orientations with respect to each other, I am still unable to
obtain a system showing h-bonds as shown below.
I have followed minimization (8000 steps), heating (20ps), equilibration
(2ns) followed by production (100-200ns, room temperature).
Is there any possible way to mimic the interactions between
two p-nitropheno molecules? It's a natural process so I believe it should
be generated using MD protocols stating from two monomers.
Thanks,
Jac
[image: Inline image 1]
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: image.png)
