Date: Fri, 1 Sep 2017 11:59:28 +0000 (UTC)
Dear all,
Currently I am working on amyloid aggregation.So I take 5 Abeta-16-22 residue peptide to show amyloid aggregation.In this case I have got different result when I am using iwrap=1 or not.My input files are : 4000ps MD NPT simulation
&cntrl
imin = 0, irest = 1, ntx = 7,
iwrap= 1,
ntb = 2, pres0 = 1.0, ntp = 1,
taup = 2.0,
ig =-1,
cut = 10.0, ntr = 0,
ntc = 2, ntf = 2,
tempi = 300.0, temp0 = 300.0,
ntt = 3, gamma_ln = 1.0,
nstlim = 2000000, dt = 0.002,
ntpr = 2000, ntwx = 2000, ntwr = 2000
/
And Another is :4000ps MD NPT simulation
&cntrl
imin = 0, irest = 1, ntx = 7,
ntb = 2, pres0 = 1.0, ntp = 1,
taup = 2.0,
ig =-1,
cut = 10.0, ntr = 0,
ntc = 2, ntf = 2,
tempi = 300.0, temp0 = 300.0,
ntt = 3, gamma_ln = 1.0,
nstlim = 2000000, dt = 0.002,
ntpr = 2000, ntwx = 2000, ntwr = 2000
/
When I use iwrap=1 from the starting of production run, in secondary structure analysis I don't get any extended Beta sheet .But when I don't use iwrap(after 72 ns I have to use iwrap=1 to run the simulation more) I get extended beta sheet.I have attached my secondary structure analysis.
Please help me out .
Thanks and regardsSaikat
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
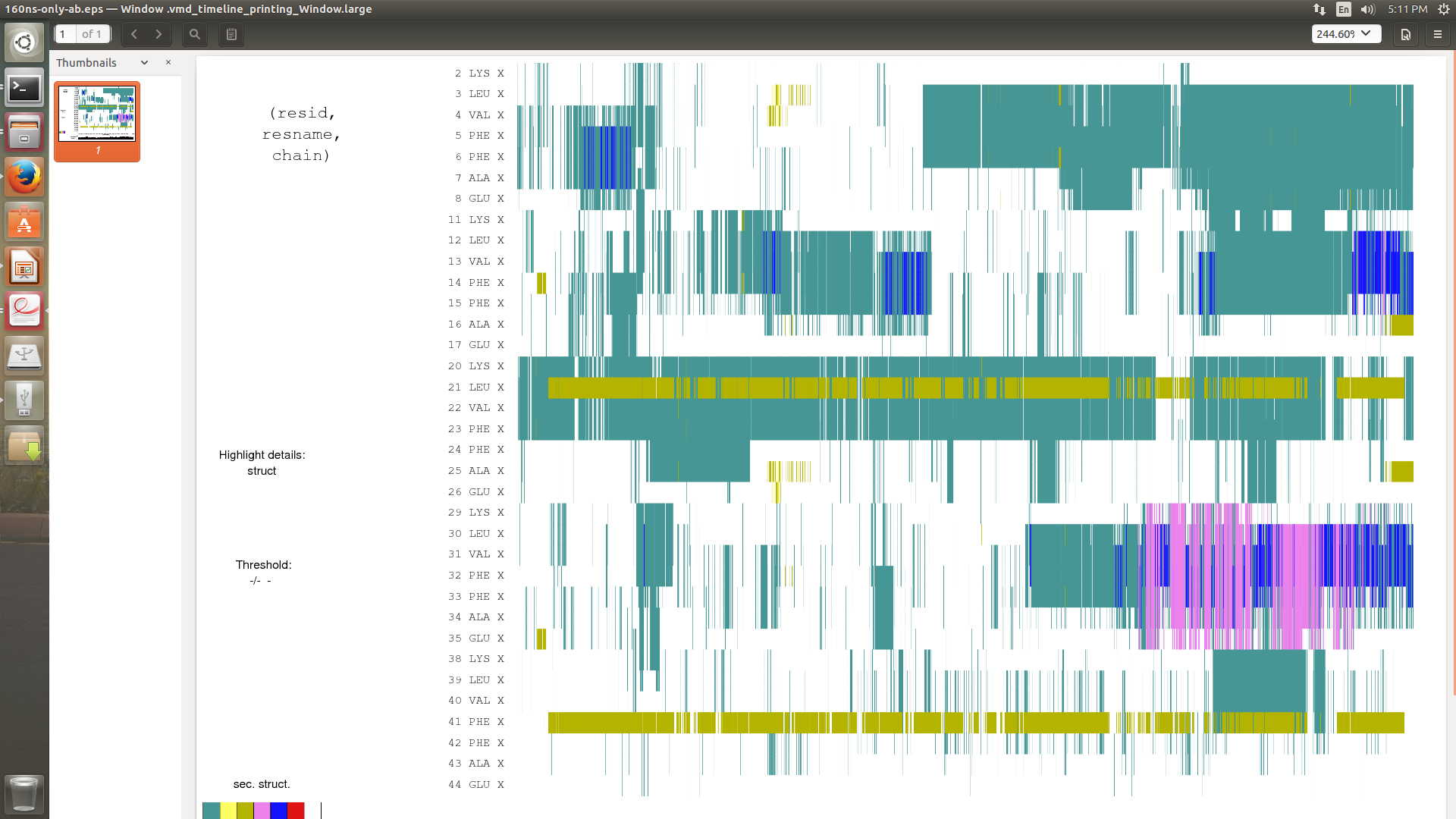
(image/png attachment: use-1wrap_1.png)
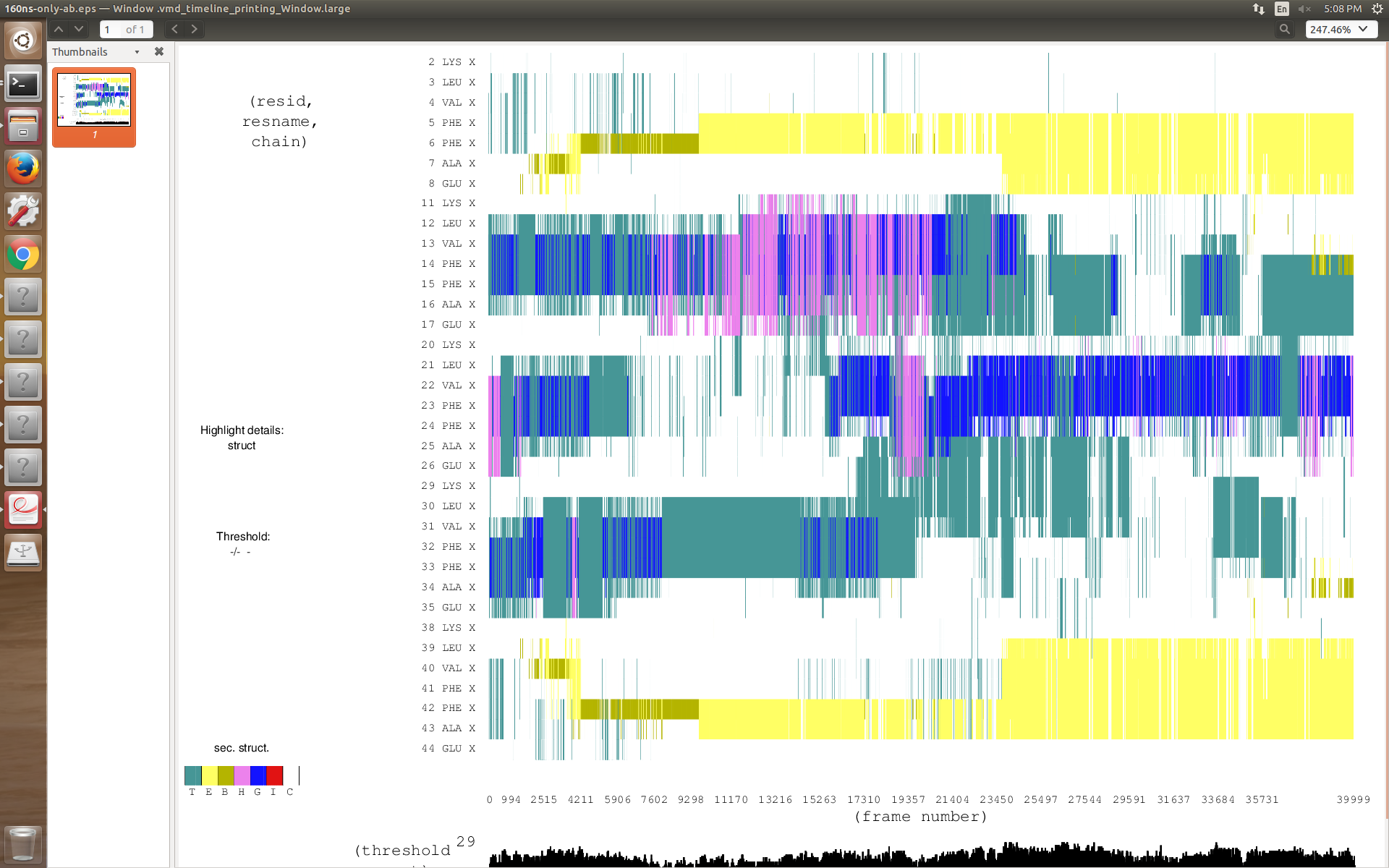
(image/png attachment: use-iwrap_0.png)
