Date: Fri, 26 May 2017 10:19:32 +0000
I guess the problem is the distortion of the tetrahedral symmetry of the carbon where the amine is attached to. Which of course is already distorted because such carbon is in a cyclopropyl, but with a simple minimization the problem gets clear. Here in attachment pictures of the initial coordinates and the minimized ones. The angles looks too distorted.
So I thought I should add parameters for the angles. But how can I do it properly? which ones to specify and how to get the correct values? I guess I should leave some bending.
Thanks,
Loris
________________________________
From: Bill Ross <ross.cgl.ucsf.edu>
Sent: Friday, May 26, 2017 10:50:10 AM
To: amber.ambermd.org
Subject: Re: [AMBER] add ligand parameters
>From a chemistry-challenged reading, it sounds like you might not be
getting planarity where you expect it?
If so, adding an improper torsion term may be the solution.
Bill
On 5/26/17 1:45 AM, Loris Moretti wrote:
> Hi All,
>
>
> I have a ligand containing a cyclopropyl substituted with an amine group and I would like to simulate its dynamic behavior inside a protein.
>
> But the parameters I get from the usual procedure antechamber + parmck result in a distorted molecule. Here in attachment the minimal example as an initial molecule (ligand.mol2) with AM1-BCC charges, the ligand.frcmod produced by the command:
>
>
> parmchk2 -i ligand.mol2 -f mol2 -o ligand.frcmod -s 2
>
>
> (from Ambertools17)
>
>
> I've also tried with parmchk (instead of parmchk2) and gaff (instead of gaff2) without improvement. The "leap" commands to produce the system and the parameters and the coordinates for the system (system_init.prm and system_init.crd). The md.in for the simulation settings and the trajectory system_md.cdf obtained with the command:
>
>
> sander -O -i md.in -o md.out -p system_init.prm -c system_init.crd -r system_md.rst -x system_md.cdf
>
>
> In the simulation is clear that the angle of the C-C-N (carbons of the cycle and the nitrogen) goes distorted and I understand I have to insert new parameters for such angle into the ligand.frcmod file. I guess I have to generate those values manually, how should I do that? I've tried to check but I couldn't find proper information about the procedure.
>
>
> Thanks in advance,
>
> Loris
>
>
>
> _______________________________________________
> AMBER mailing list
> AMBER.ambermd.org
> http://lists.ambermd.org/mailman/listinfo/amber
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
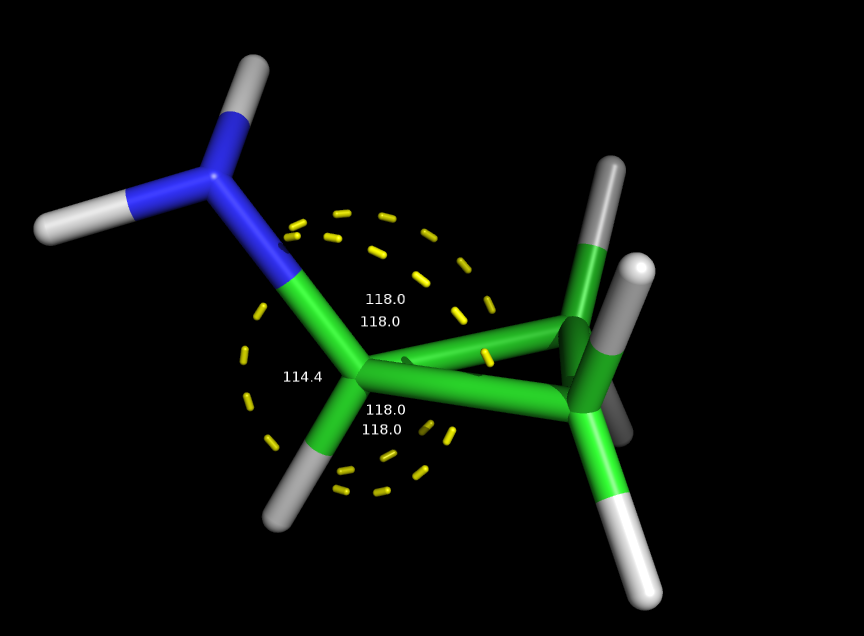
(image/png attachment: system_init.png)
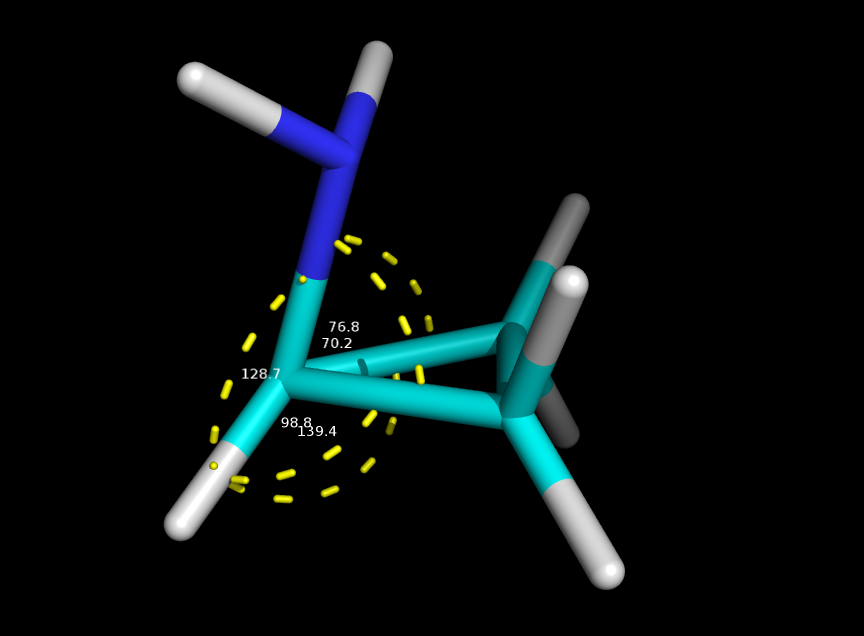
(image/png attachment: system_min.png)
