Date: Mon, 14 Nov 2016 22:29:17 +0000
________________________________
Fra: Oliver Emil Skytte Glue
Sendt: 14. november 2016 23:24
Til: amber.ambermd.org
Emne: Positive interaction energy between Sodium ions?
Hi,
I've been simulating the Dickerson dodecamer(CGCGAATTCGCG) with AMBER 14 and 16 on either GTX 1080 or Tesla K20 with PMEMD.CUDA in the NPT ensemble with the Langevin thermostate and the Monte Carlo barostate.
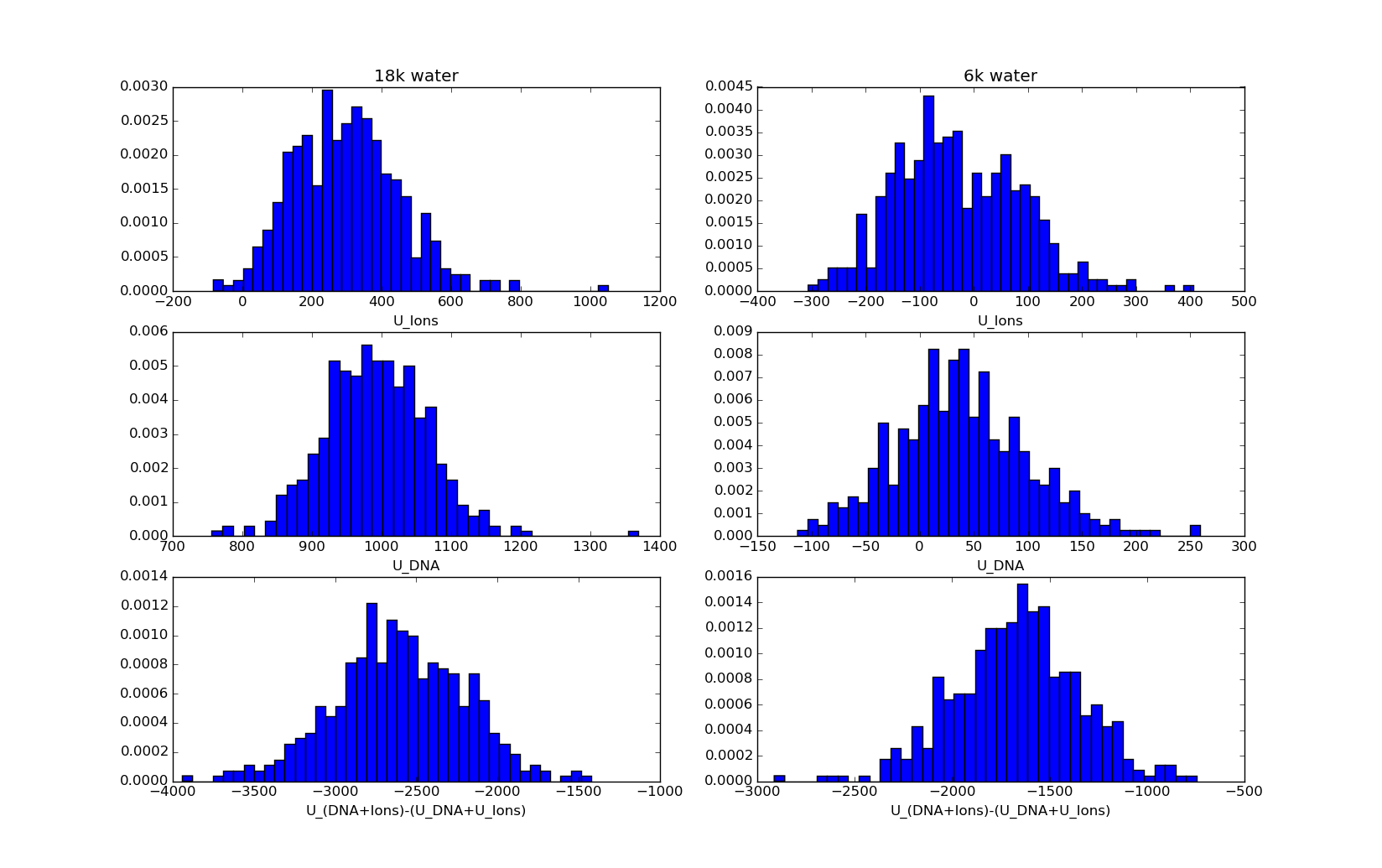
Consider two systems of the DNA neutralized with 22 sodium ions each and solvated with 6k and 18k water molecules(OPC) simulated for 1 Ás+. For the 'small' 6K water system, the electrostatic interactons of sodium - sodium fluctuate around 0 kcal/mol with a standard deviation of ~200(calculated with either imin=5 or esander by stripping water and DNA).
For the 'big' 18k water system, the electrosatic interactions are above 0 for 99.9+% of the frames - I guess within the accuracy of PME?
How is it possible for the sodium sodium interactions for the small system to be negative?
This further gives raise to different electrostatic energies of the DNA of the two systems and interaction energies between the DNA and Ions. I've attached a picture showing histograms of the electrostatic potential energy for the two systems.
The histograms doesn't look very nice as only 500 frames were calculated from microsecond simulations.
Any tips? This seems wrong and unphysical!
I can also provide more details.
Many Thanks,
Oliver Glue, University of Southern Denmark
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: electrostatic_energy_-_DNA__Ions.png)
