Date: Tue, 27 Sep 2016 11:22:41 +0000 (UTC)
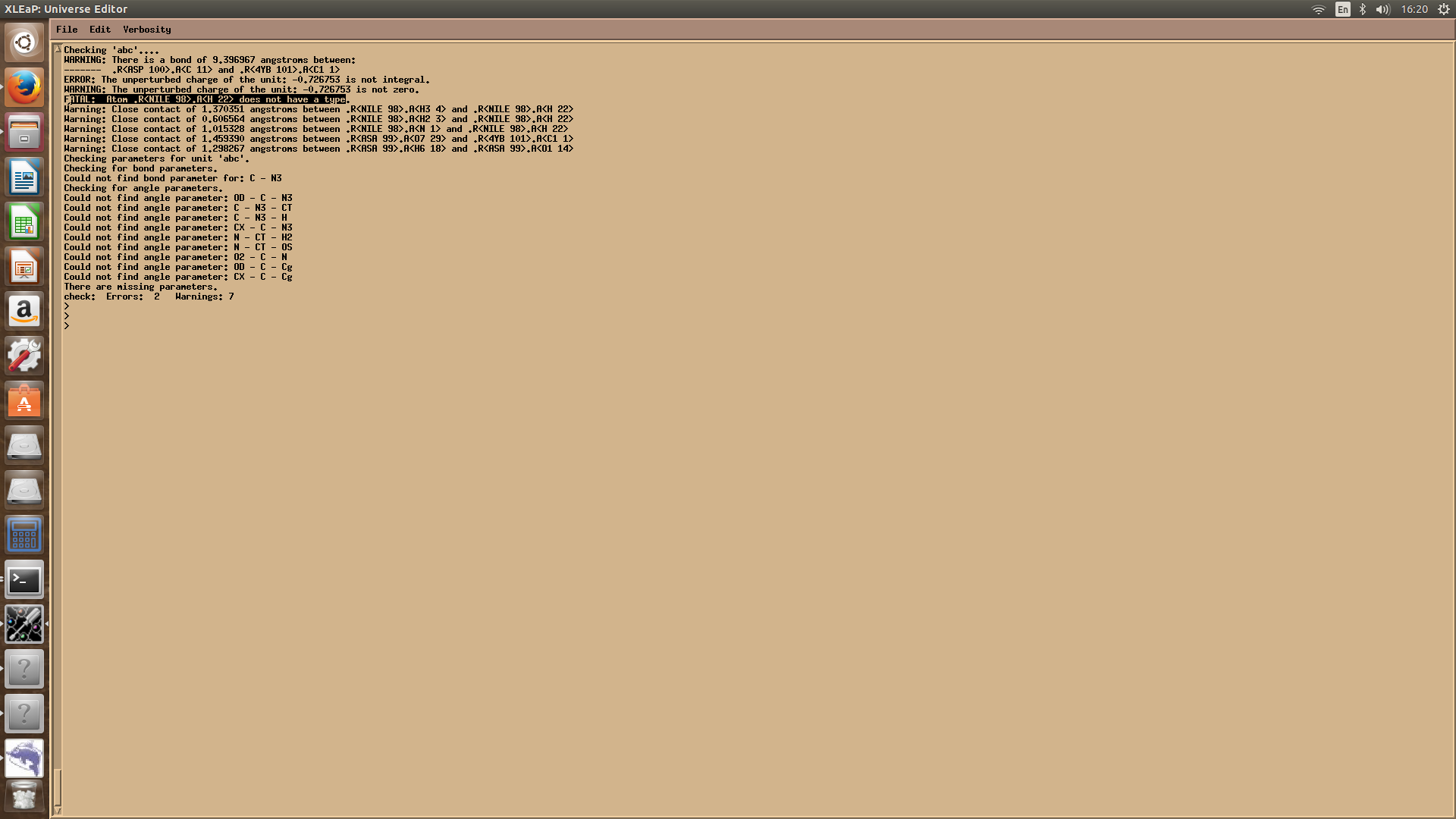
Hi all,I am currently trying to simulate a non standard amino acid i.e. N-glycosylation. I have generated parameters for this non standard residue and loaded other libraries for protein and glycan in xleap but it still gives some error messages which I could not understand why its happening, although every residue is parametrized.
Image of the error message is attached. Any help is highly appreciated.
Regards,Mahrukh
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: Screenshot_from_2016-09-27_16:20:24.png)
