Date: Fri, 20 Feb 2015 14:46:52 +0100
I would like to also try the constraints in sander to fold the ssDNA.
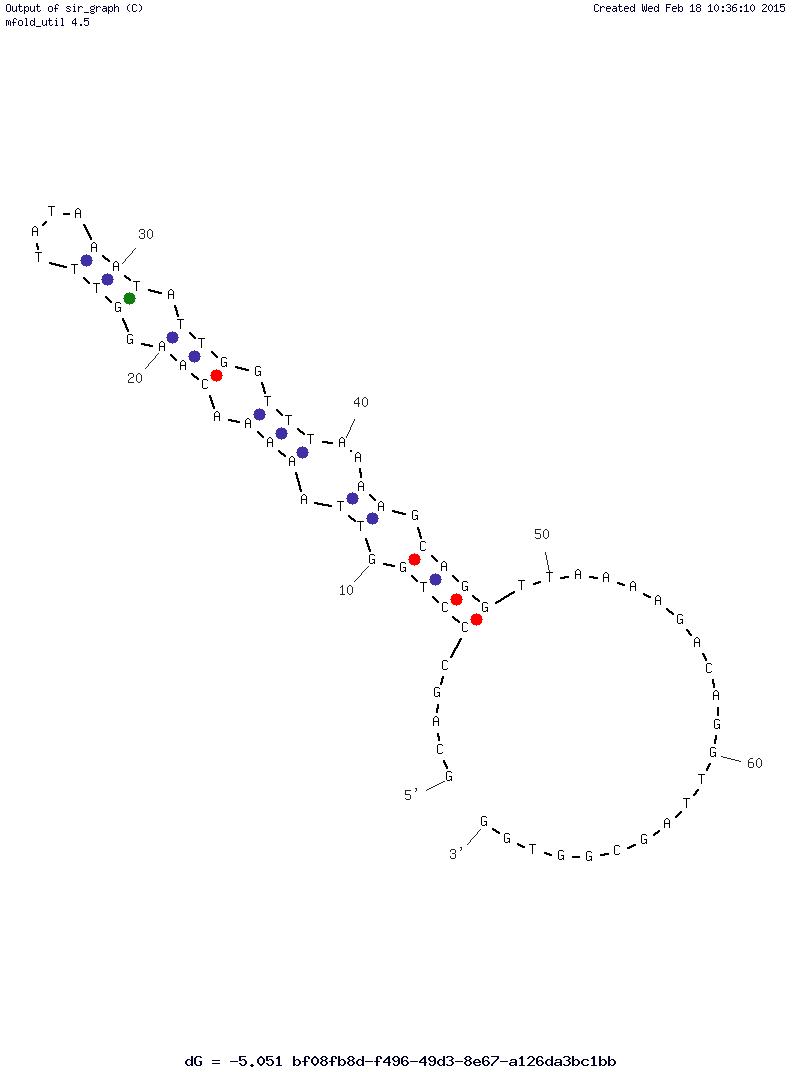
Which constraints should be the best for this case? I have attached the 2D
ssDNA conformation that I would like to get.
best regards
Urszula
> On Thu, Feb 12, 2015, Urszula Uciechowska wrote:
>>
>> I was wondering if its possible to build a ssDNA in hairpin conformation
>> using nab? Does anyone know other software where I could do this?
>
> It is certainly *possible*: the pseudoknot example could be adapted to a
> hairpin. It uses distance geometry to construct conformations satisfying
> certain restraints, such as WC pairing in the stem.
>
> Having said that, there are lots of other solutions, and a Google search
> will
> point to many of them. I personally would use CYANA for this purpose, but
> that is largely because I'm familiar with that code. You could also set
> up
> constraints in sander and "fold" the dna from an extended configuration.
>
> Maybe others on the list will have more specific suggestions.
>
> ....dac
>
>
> _______________________________________________
> AMBER mailing list
> AMBER.ambermd.org
> http://lists.ambermd.org/mailman/listinfo/amber
>
>
University of Gdansk and Medical Univesity of Gdansk
Department of Molecular and Cellular Biology
ul. Kladki 24
80-822 Gdansk
Poland
-----------------------------------------
Ta wiadomość została wysłana z serwera Uniwersytetu Gdańskiego
http://www.ug.edu.pl/
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/jpeg attachment: ssDNA-hairpin.jpeg)
