Date: Wed, 5 Mar 2014 19:18:48 +0530
Dear Prof Case and Jason
Thanks for the prompt reply
I am using VMD for visualizing.
1) I start VMD
2) New Molecule -> ADNA.top (leap generated) (format:Amber7parm)
3) Load data into molecule -> ADNA.crd (leap generated) (format: Amber
coordinates with periodic box)
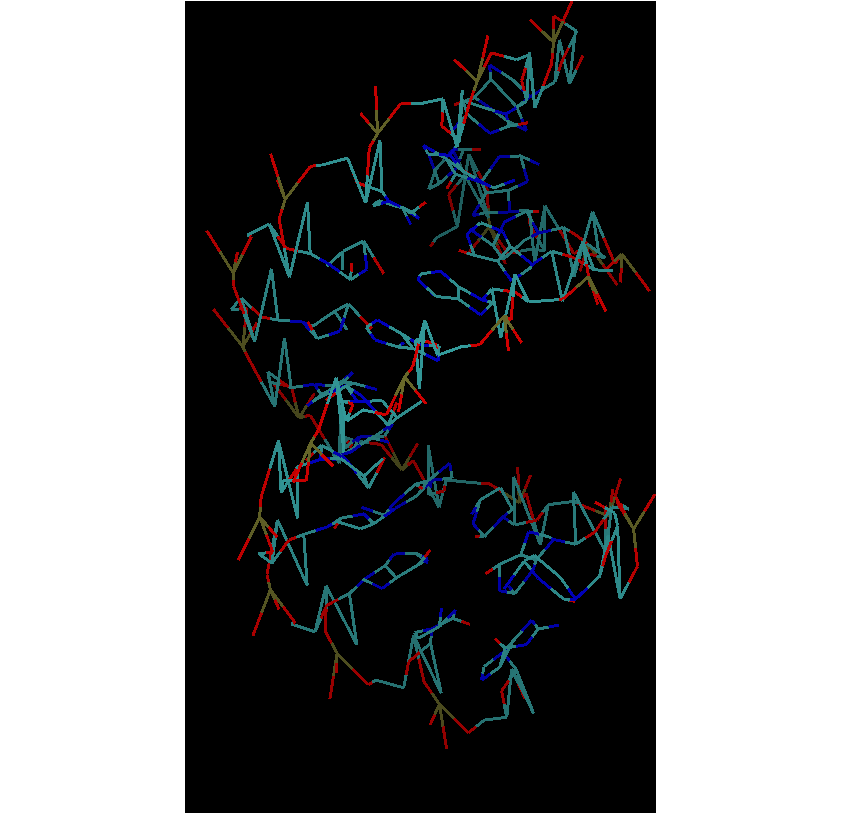
And the entire structure appears distorted. Sorry for the confusion. The
structure is distorted at this stage itself. I have attached a snapshot. I
did not use solvateBox. I packed water with packmol and used the set
command for dimensions.
As suggested by Jason, I checked the restraint energy in the 1st step of
minimization, it is 70.73kcal/mol (the rk2 value I gave was 100.0, I know
its kind of high but I was just changing the force constant to see if
things improve before I figured out the starting structure itself was
distorted). There are no overlaps either as checked with the mentioned
command.
Thanks
On Wed, Mar 5, 2014 at 6:47 PM, David A Case <case.biomaps.rutgers.edu>wrote:
> On Wed, Mar 05, 2014, DEBOSTUTI GHOSHDASTIDAR wrote:
> >
> > I went a step back and visualized the leap
> > generated pdb and it was a perfect A-DNA. However, when I loaded the leap
> > generated crd file on the leap generated topology file, the structure
> > looked severely distorted.
>
> This should not happen. The coordinates leap puts into the pdb file
> should be the same as those it puts into the crd file. (If you are using
> solvateBox, add "set default nocenter on" at the top of your leap.in
> file.)
>
> It sounds like the problem is at the LEaP step: if you start with a
> severely
> distorted structure, you are indeed likely to have problems. You need to
> try
> to figure out what is going wrong at this early step.
>
> > I guess the A-DNA structure was disturbed during minimization.
>
> This is confusing: above you said you were seeing distortions with the
> "leap generated crd file on the leap generated topology file". If so, bad
> things must have happened before any minimzation.
>
> >
> > For re-confirming, I visulalized the leap generated crd and topology
> files
> > given in the Amber tutorials Section B1 (Simulation of DNA) and even that
> > structure was distorted. Where am I going wrong?
>
> What visualization program are you using? Exactly what commands are you
> executing? The problem may be in how you are using the visualization
> program,
> but we can't help there, since we don't even know which program you are
> using.
>
> ...dac
>
>
> _______________________________________________
> AMBER mailing list
> AMBER.ambermd.org
> http://lists.ambermd.org/mailman/listinfo/amber
>
-- Debostuti
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: ADNA_leap.png)
