Date: Mon, 01 Jul 2013 16:20:54 +0200
Dear all,
I am just curious if there is any recommended approach
how to create periodic box around solvated (externally)
lipid bilayer system.
In my case I first equilibrated membrane just with 20A of water
on both sides (added via packmol). The periodic box I created
using tleap command "setbox UNIT vdw". However this approach is not
ideal and in fact on sides one obtain some "vacuum" layers due to
the "random" orientation of lipid aliphatic tails on the side box surface.
This effect is not so problematic if the water layer is not too high and
can be equilibrated after some relatively short NPT run. However if one
adds around 80A of water
there is problem and density equilibration can take significant time.
I wanted to prevent me from such problems so when I constructed my final
system
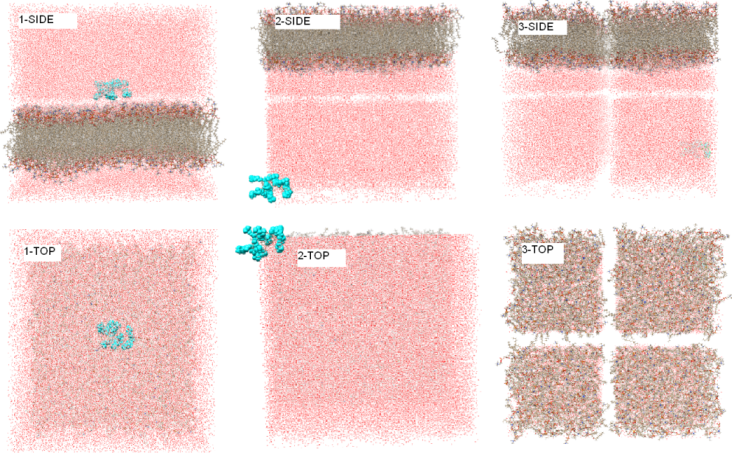
= previously equilibrated lipid bilayer + ligand (cyan in attached figure)
+
water (20A bottom), (80A top) - see the part 1-SIDE/TOP of the attached
figure,
I tried to construct my box using the X, Y dimensions written in *.rst
file of the equilibrated
membrane which I used to build the whole complex and dimension in Z
direction (of the whole complex)
I obtained using "setbox UNIT vdw" (I used "set UNIT box { X Y Z }"
command in the final step).
This approach unfortunately failed and starting the heating (NVT) phase
the system was reimaged in some strange unwanted way (see 2-SIDE/TOP in
attached figure). Then I tried also some X, Y dimensions between those
"vdw" and those obtained from *.rst of the membrane equilibration run
(e.g. average), but it creates even more bizarre system reorganization -
see the attached figure part 3.
Let's just notice that during the minimization phase the system was
maintained in a good
box, the problem with wrong reimaging starts from the NVT MD.
So to prevent from really significant equilibration phase (of course with
constrained lipids) I can
imagine that perhaps might help to create water layers which in X and y
direction agree with dimensions defined by lipid tails (rather than lipid
heads). But anyway then I will have
using "setbox UNIT vdw" perfectly continuous infinite layers of water but
partially isolated pieces of lipid blocks, which maybe connect after some
significant time of simulation so even this idea is
not the optimal one.
So my question is which is actually the best (recommended) approach to
create periodic
box around solvated lipid bilayer systems to prevent any discontinuity on
the start,
e.g. to obtain perfectly following-up (lipid part and water part) periodic
images ?
I also wonder why does not work the approach with X, Y dimensions taken
from the *.rst file
of the equilibrated membrane which was used for the final complex ? I
thought that this
has to be the most optimal option and for sure that it has to work
especially if the additional
water layers was constructed to be in the X, Y intervals of the original
"heds solvating" water
molecules from the previous just membrane equilibration run.
Thanks a lot in advance for any comments !
Best wishes,
Marek
-- Tato zpráva byla vytvořena převratným poštovním klientem Opery: http://www.opera.com/mail/
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: boxes_fin.png)
