Date: Fri, 22 Feb 2013 18:10:20 +0000
Hi all,
I'm running a long MD simulation of 100ns of a ligand bound to a protein
using amber 10. I turned iwrap=1 to avoid the simulation stalling due
to precision errors.
##################
production run infile
&cntrl
nstlim=2000000, dt=0.002, ntx=5, irest=1, ntpr=500, ntwr=500, ntwx=500,
temp0=300.0, ntt=1, tautp=2.0, iwrap=1, ig=-1
ntb=2, ntp=1,
ntc=2, ntf=2,
nrespa=1,
&end
#################
I ran a quick test measuring the distance from the hydroxyl group of the
ligand which a primary alcohol with a cis-3-4 double bond to Tyr120. The
distances seemed quite large (distance graphs for one of the ligands
attatched). When I check the trajectories with vmd or chimera, the ligand
does not seem to be behaving properly: either coming out of the pocket or
getting stuck on part of the protein (when visualized with vmd).
I'm pretty sure this is an imaging problem. I have more simulations where
the ligand comes out of the pocket for a some frames then goes back in. the
larger image enclosed illustrates the issue across 3 repeats of 4 different
ligands bound to the same protein. Each graph measures distance between the
ligand and Tyr120 of a different trajectory. The protein MUP is like a
barrel and the ligand is expected to be relatively mobile within the
pocket, but prob not to this extent.
I have looked through the archives and tried applying the following ptraj
imaging commands with no success;
The protein residues = 1-157
The ligand = 158
#############
trajin md_1.crd
center :1-157 mass origin
image :1-157 origin center bymask :1-157 familiar
center :1-158 mass origin
image :1-158 origin center bymask :1-158 familiar
trajout md_1_manImaged.ncdf netcdf
#############
Zipped Files containing the first 300 frames of a original (before imaging)
trajectory and topology files can be downloaded from the following links:
1) 3c6-ligand-protein:
https://dl.dropbox.com/u/12686110/04_3c6.zip
Any help anyone could offer to solve this issue would be greatly
appreciated.
Best regards,
ET
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
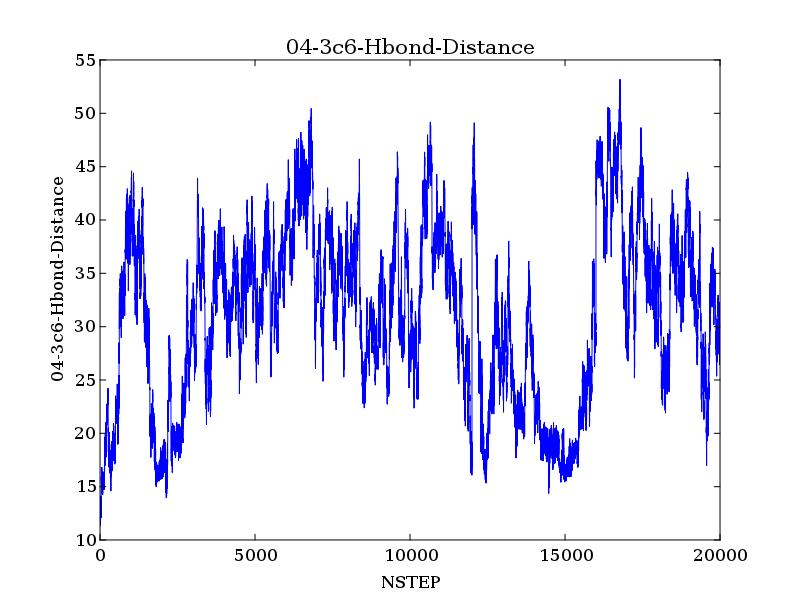
(image/jpeg attachment: 04-3c6-Hbond-Distance.jpg)
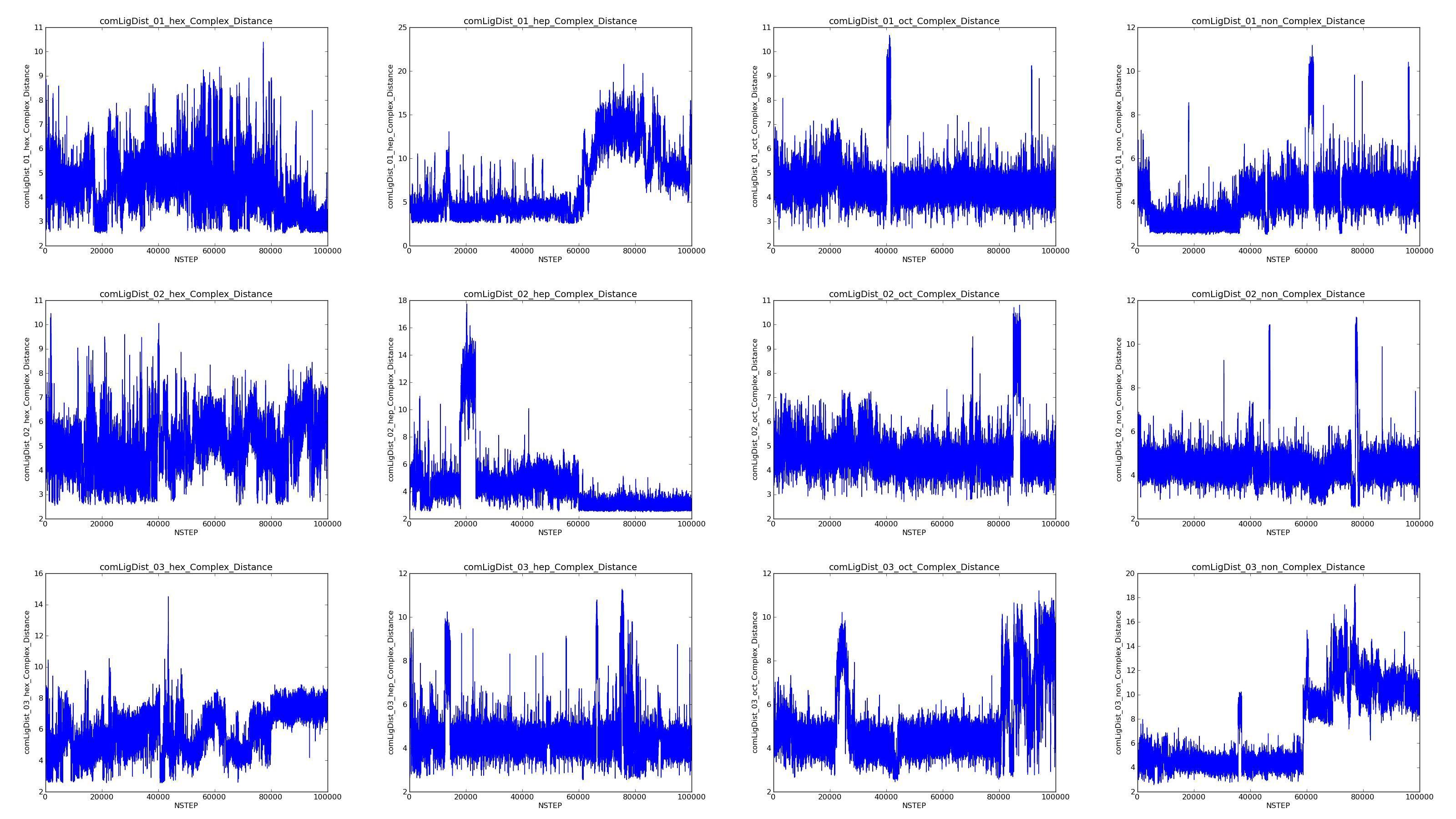
(image/jpeg attachment: Prim-ols_RAWComplex_DistanceFin.jpg)
