Date: Fri, 1 Feb 2013 11:31:55 +0100
Dear Amber users,
I am trying to get an average structure, using the the following script in Ptraj:
trajin PROD6.mdcrd
center :1-445 mass origin
image origin center
rms first mass out rms.dat :1-445
average model-1_average.pdb pdb
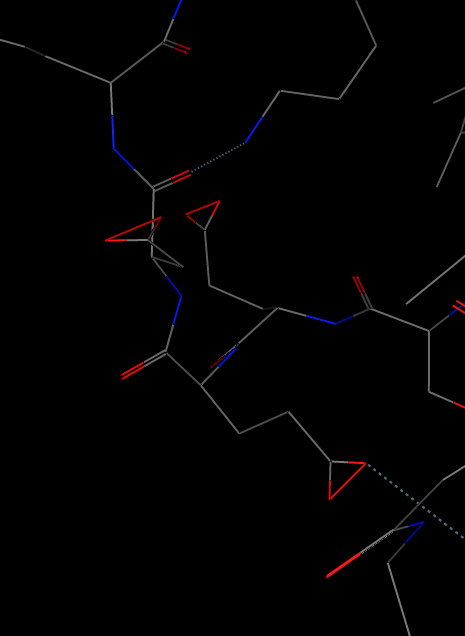
But in the average structure I got strange connections between oxygen atoms, please see attachment.
Could anyone help me to figure out what went wrong with the calculation?
How should I modify the script?
/Urszula
--------------------------------------------------------------
Urszula Uciechowska, Ph.D.
Department of Chemistry
Umeň University
SE-901 87 Umeň, Sweden
urszula.uciechowska.chem.umu.se
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: average.png)
