Date: Sat, 15 Dec 2012 12:25:15 -0800 (PST)
Dear All,
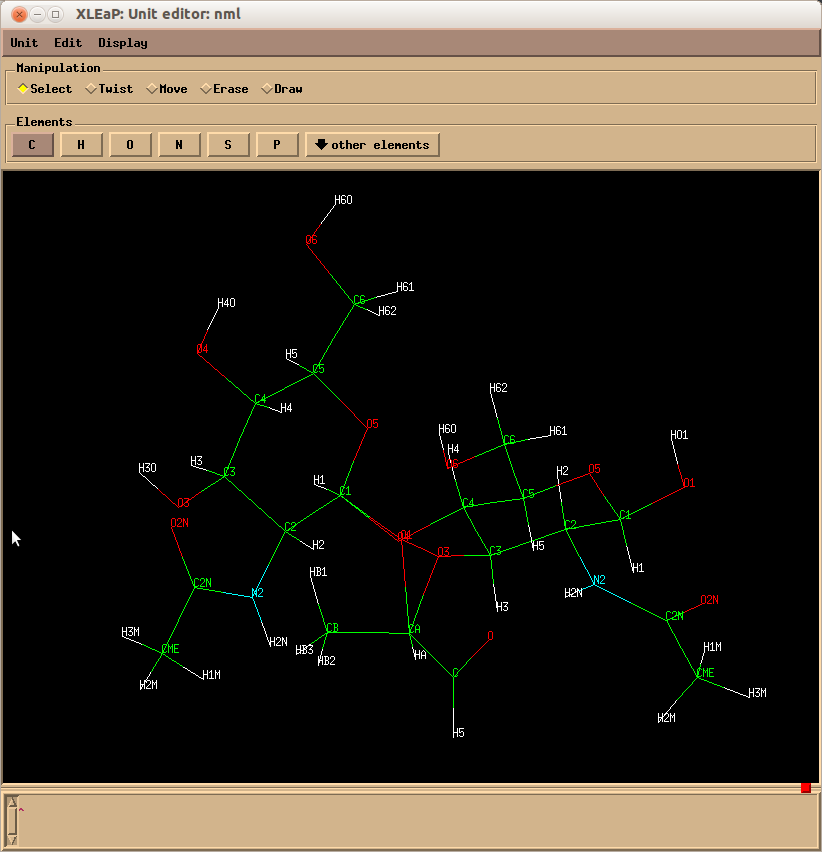
I am trying to generate a disaccharide consisting bGlcpNAc[1,4]aGlcpNAc[3-]-lactic acid in leap, see in the attched pdb file.
Could someone suggest me the monosaccharide units I should use for
this molecule? My own attempt from 4 residues ROH 1, WYB 2, 1YB 3 and
a self-constructed LAC 4 residue has incorrect connections, please, see the screenshot of molecule in the editor.
GlcpNAc[3-]-lactic acid is in fact muramic
acid, but I did not see separate PDB code for in the GLYCMA06.prep
file, so I made it from GlcpNAc and a lib file for lactic acid in
antechamber with gaff atomtypes. Is there a better way for muramic acid?
Thanks in advance,
Krisztina
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
- X-unknown/x-pdb attachment: NML2.pdb
(image/png attachment: NMLwithGAffBasedLAC.png)
