Date: Wed, 21 Nov 2012 15:19:15 +0530
Hi all,
I am running a minimization using sander on some ligand-protein complex..
but when I retrieve the PDB file of minimized structure, ligand appears
broken as seen in attached pictures..
I had used gaussian on the ligand and used the gaussian output as input for
antechamber..
My input file is:
&cntrl
imin=1, maxcyc=7500, ncyc=2500,
ntpr=5,ntr=1,restraint_wt=10, restraintmask=':1-356',
cut=16, ntb=1,
/
what could be the reason? How can I rectify this?
-- Jitesh V. Doshi Research Associate, Bioinformatics Centre, University of Pune.
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
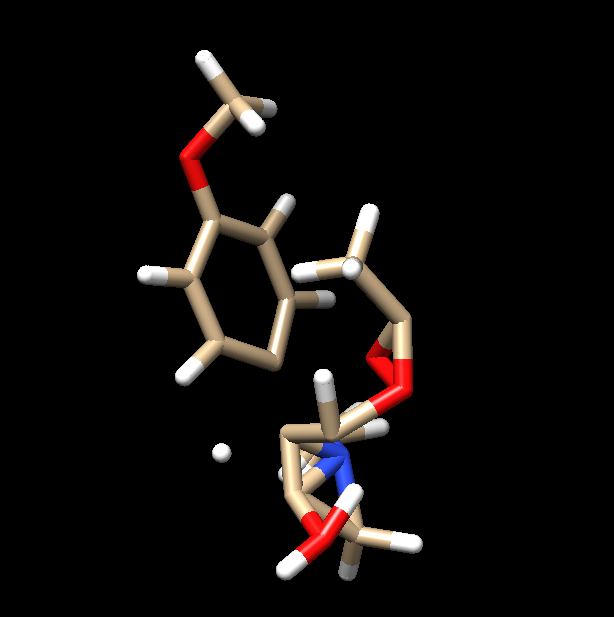
(image/png attachment: lig_after_min.png)
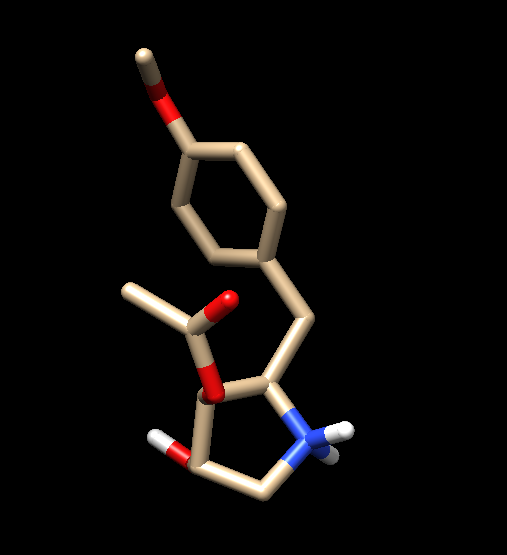
(image/png attachment: lig_before_min.png)
