Date: Sat, 1 Sep 2012 18:40:07 +0200
Friends,
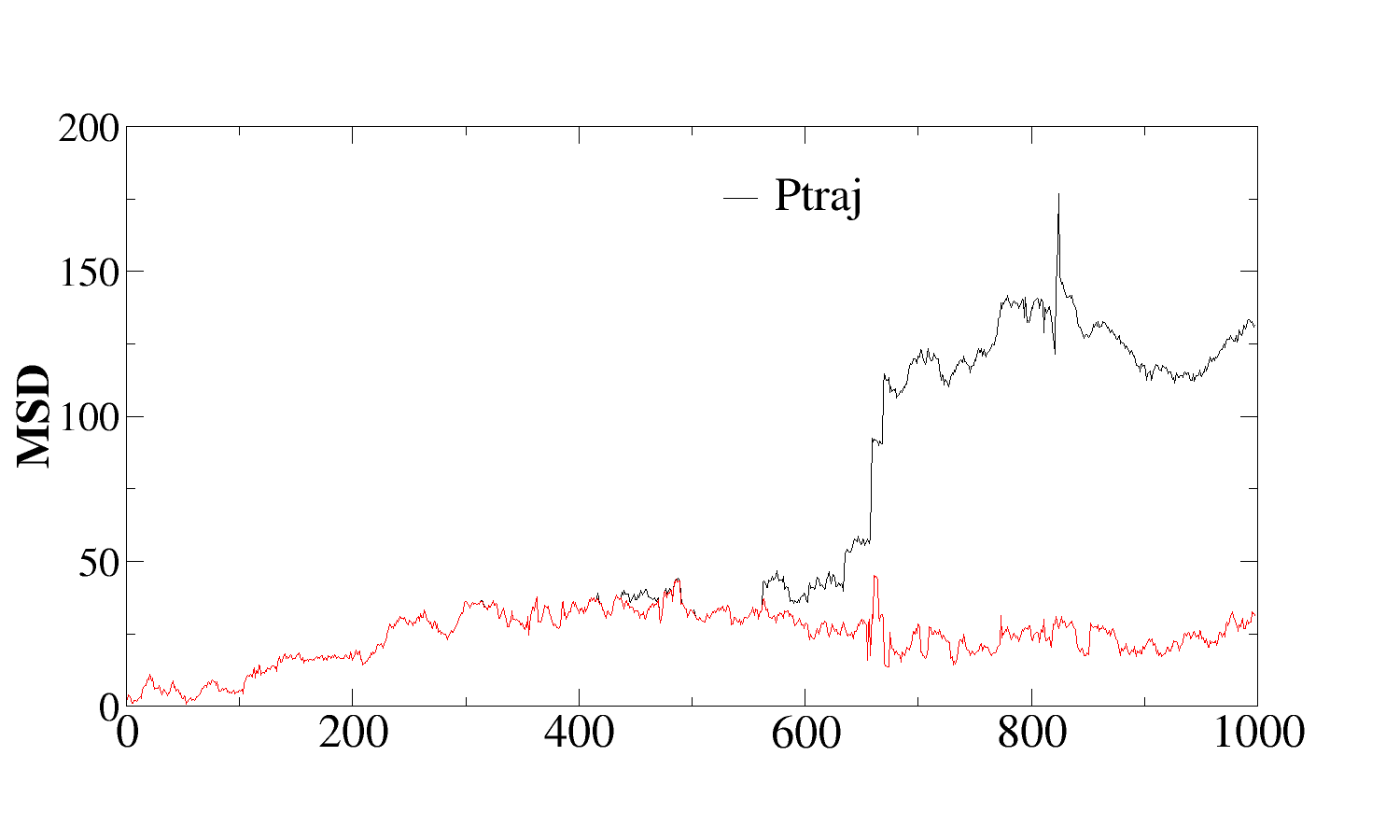
I calculated the msd of a specific water (residue 3457) oxygen by an
in-house script and compared with ptraj. The values were same and
diverged at a point (frame 565 onwards).
I checked the diffusion routine within actions.c and found that the
coordinate is corrected for periodicity as follows ( lines7126-7138)
if delta (current – prev) > box/2.0: delta = delta – box
if delta (current - prev) < - box/2.0: delta = delta + box
If I just compare the distance of the oxygen atom from frame 565
(f565.pdb attached) to frame 1 (reference.pdb), the delta values for
x,y,z are:
dx = (1.959-24.629) = -22.670 and sq. of it = 513.93
dy = (5.701-30.283) = -24.582 and sq. of it = 604.27
dz = (30.053-44.725) = -14.672 and sq. of it = 215.27
Now if you see the attached x,y,_z output of ptraj. The sq of dy is
1075.85. My box size is as follows: 57.73 57.38 57.75.
I understand that ptraj add box size to dy = -24.582 + 57.38 = 32.798
and sq. of it = 1075.85.
But I dnt understand why does ptraj do the addition of box size when
either of the conditions for periodicity correction is not is “true”
in this case. Someone please write me if there is something wrong with
ptraj or my understanding.
On Fri, Aug 31, 2012 at 7:47 PM, Bala subramanian
<bala.biophysics.gmail.com> wrote:
> Hi,
> Thank you for the details. I have written small code to calculate msd
> and compared with ptraj for a single water oxygen. I get the same
> values upto a point and then a divergence (attached fig). I dnt know
> if this could be due to periodicity problem.
>
> I would like to know how periodicity is taken care of while
> calculating the msd. Could someone please guide me on the same.
>
> thanks,
> Bala
>
> On Sat, Aug 25, 2012 at 5:59 PM, Dmitry Mukha <dvmukha.gmail.com> wrote:
>> Hi All,
>>
>> 2012/8/23 Daniel Roe <daniel.r.roe.gmail.com>:
>>> Hi,
>>>
>>> >
>>> More specifically, in the "<file>_a.xmgr" file the second column is
>>> the average distance travelled by all the atoms, i.e. sqrt(
>>> SUM[dist^2]/N ), where N is the number of atoms. Columns 3 and 4 in
>>
>> Actually, you are not right. sqrt(SUM[dist^2]/N )>=SUM[dist]/N, and a
>> case of equality appears when 'dist'=const.
>>
>> In general form, for large numbers, mean square = square of mean + variance
>>
>> 2Bala:
>>
>> If you use 'average' key word, you will get only 2 columns, one for
>> time and one for the appropriate mean value.
>> _r - mean square displacement,
>> _a - mean displacement,
>> _x, _y, _z - Cartesian decomposition.
>>
>>> this case are the distance travelled by each individual atom, i.e.
>>> sqrt( dist^2 ), so col2 should not be equal to the straight average of
>>> cols 3 and 4, but rather col2 = sqrt( (col3^2 + col4^2) / 2), which
>>> you can verify it is.
>>>
>>> Hope this helped.
>>>
>>> -Dan
>>>
>>> --
>>
>> --
>> Sincerely,
>> Dmitry Mukha
>> Institute of Bioorganic Chemistry, NAS, Minsk, Belarus
>>
>> _______________________________________________
>> AMBER mailing list
>> AMBER.ambermd.org
>> http://lists.ambermd.org/mailman/listinfo/amber
>
>
>
> --
> C. Balasubramanian
-- C. Balasubramanian
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: MDA.png)
- chemical/x-pdb attachment: f565.pdb
- chemical/x-pdb attachment: reference.pdb
- application/octet-stream attachment: watdif_x.xmgr
- application/octet-stream attachment: watdif_y.xmgr
- application/octet-stream attachment: watdif_z.xmgr
