Date: Tue, 17 Jan 2012 18:42:51 +0530
Dear All AMBER users,
I performed MD simulation using pmem.cuda (AMBER 2011 package) on GPU
workstation and found that trajectory does not stabilized even after 10ns.
the details are as follows :
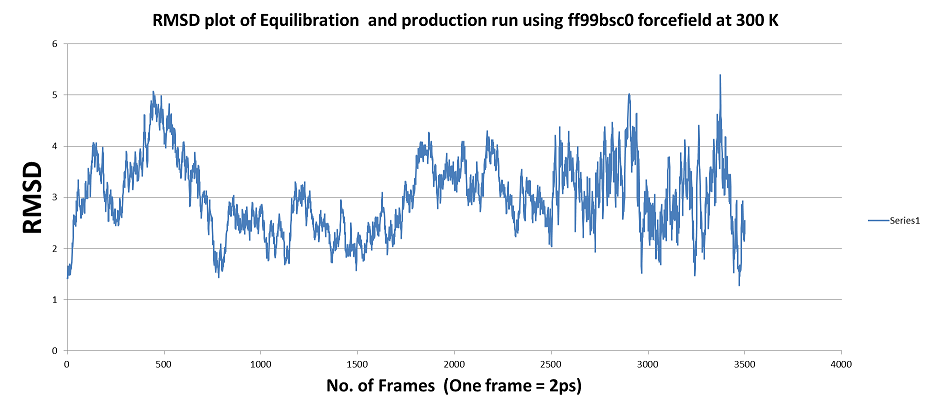
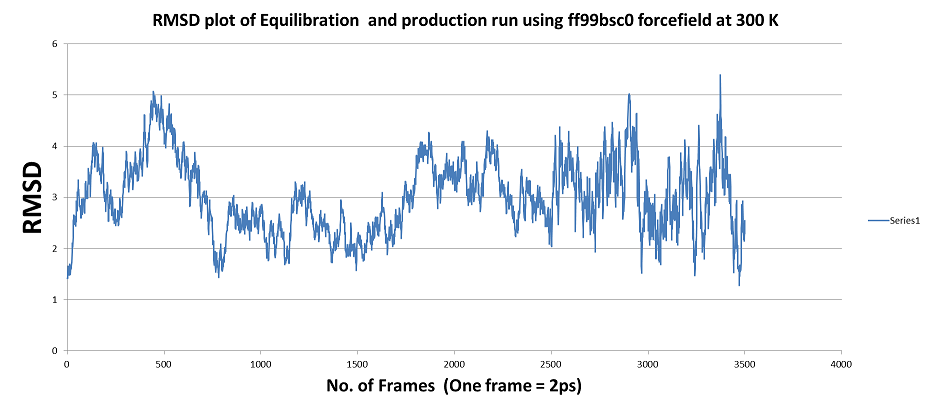
1) MD simulation using ff99bsc0 forcefield: even after ~12ns of MD run
the trajectory is not stabilized (TRAJECTORY SHOWN BELOW ) ???
[image: QJF_ff99bsc0.png]
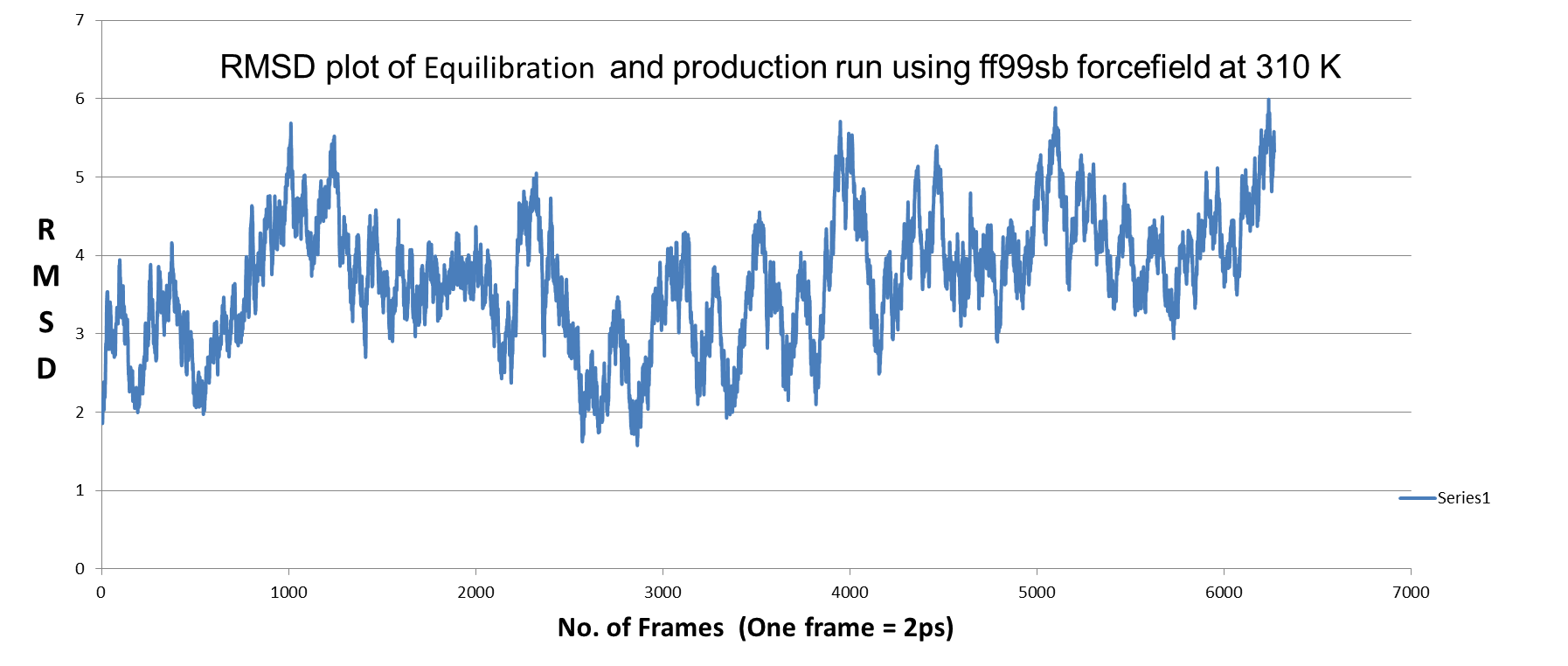
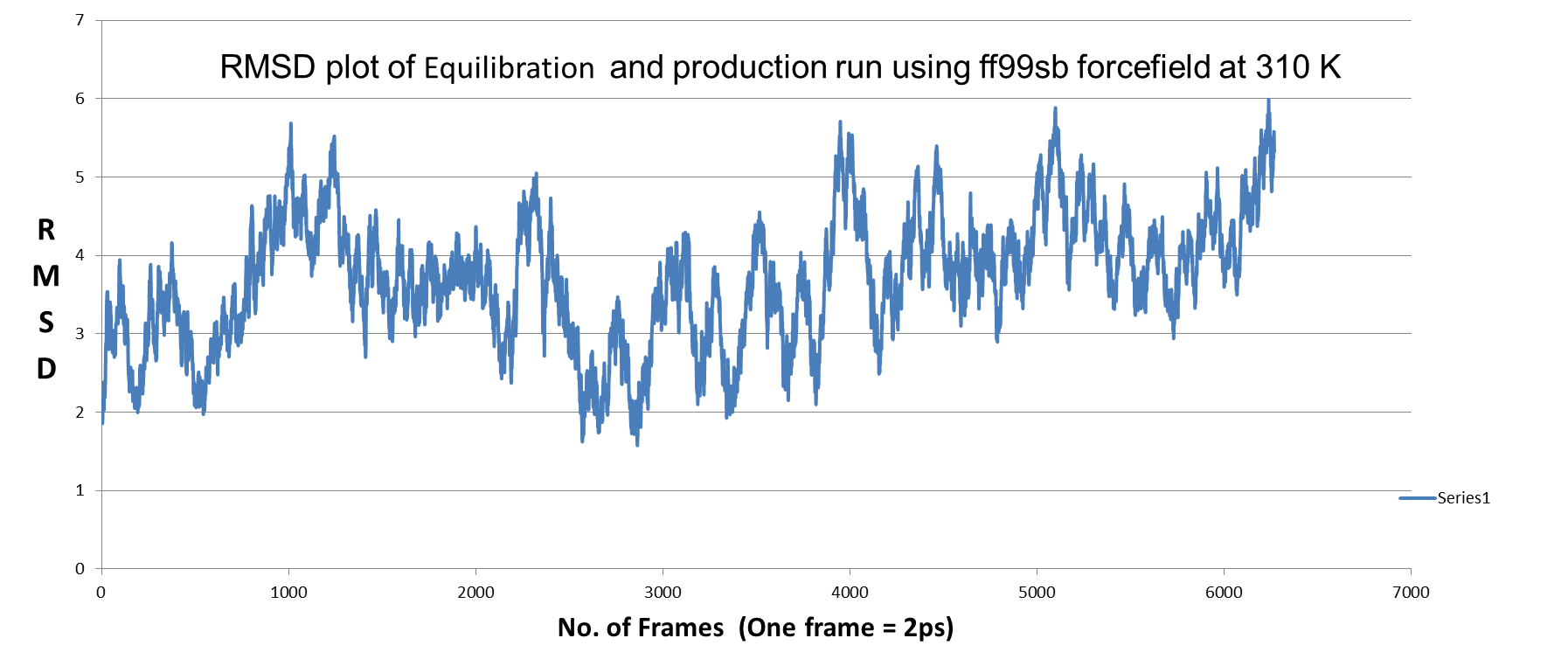
2) MD simulation using FF99SB force field: in this case also trajectory not
stabilized (RMSD plot below) ??
[image: QNF_ff99sb.png]
I am also attaching images (above) and input files for MD runs
Thanking you
Waiting for your reply..
-- -- Tumbi Muhammed Khaled Abdul Waheed, Research Scholar, Department Of Pharmacoinformatics, NIPER, S.A.S. Nagar, Mohali, Punjab, India. Mob. +91 78145 24855 www.niper.ac.in
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: QNF_ff99sb.png)
(image/png attachment: QJF_ff99bsc0.png)
(image/png attachment: 03-QJF_ff99bsc0.png)
(image/png attachment: 04-QNF_ff99sb.png)
- application/octet-stream attachment: density.in
- application/octet-stream attachment: equil-1.in
- application/octet-stream attachment: heat.in
- application/octet-stream attachment: min1.in
- application/octet-stream attachment: min2.in
- application/octet-stream attachment: production.in
