Date: Tue, 4 Oct 2011 15:35:03 -0500
Dear amberers,
I have successfully created a solvent box containing many solvent
molecules using packmol.
In xleap I used:
setbox benbox vdw 2
To generate the periodic box, and it looks fine using:
Edit benbox
But upon loading the prmtop (Amber7 format) and the crd file (Amber
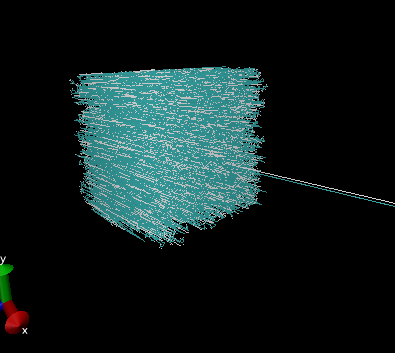
coordinates with periodic box) into VMD
the connections are all messed up. (See attached file)
Any hints?
The last few lines of the periodic .crd file are:
41.2660000 29.6810000 44.4370000 41.2710000 28.8110000 45.5290000
42.1650000 29.0160000 46.5820000 43.0540000 30.0920000 46.5430000
43.7420000 31.8010000 45.4190000 42.1510000 31.4350000 43.5470000
40.5690000 29.5210000 43.6170000 40.5780000 27.9730000 45.5600000
42.1680000 28.3380000 47.4330000 43.7500000 30.2520000 47.3620000
103.8160000 103.5940000 103.8160000 90.0000000 90.0000000 90.0000000
Thanks in advance!!
DeanDr. Dean Cuebas, Associate Professor of Chemistry
deancuebas.missouristate.edu, Ph 417-836-8567 FAX 417-836-5507
Dept. of Chemistry, Missouri State University
Springfield, Missouri 65897
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: Screen_shot_2011-10-04_at_3.33.08_PM.png)
