Date: Fri, 23 Sep 2011 16:15:37 +0700
Dear All,
I have been trying to calculate Free Binding energy of my system ( 260
residues and 1 ligand, 20 nanoseconds of MD simulation). However, I notice
that the results are weird. I don't know whether the problem arises from my
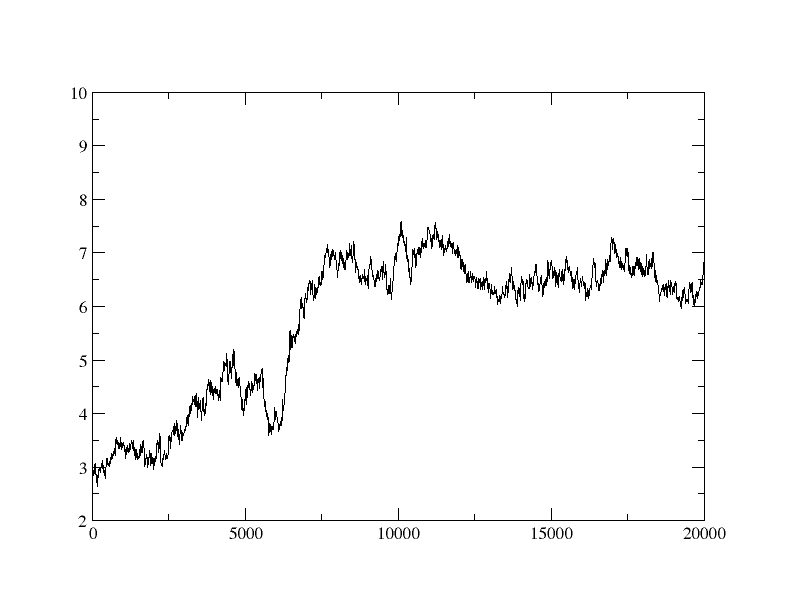
system or not. Attached here is graph of rmsd. input and output files are
described as follows. Please give me some advice if you has encountered
such a problem before. Thank you very much in advance.
My output
|Input file:
|--------------------------------------------------------------
|mmpbsa
|&general
| endframe=100, verbose=1,
|/
|&pb
| istrng=0.100,
|/
|&nmode
|nmstartframe=5, nmendframe=20, nminterval=2,
|maxcyc=50000, drms=0.4,
|/
|--------------------------------------------------------------
|Solvated complex topology file: topol/complex.prmtop
|Complex topology file: topol/com.prmtop
|Receptor topology file: topol/rec.prmtop
|Ligand topology file: topol/ligand.prmtop
|Initial mdcrd(s): prod4.mdcrd
|
|Best guess for receptor mask: ":1-269"
|Best guess for ligand mask: ":270"
|Ligand residue name is "SUS"
|
|Calculations performed using 100 frames.
|NMODE calculations performed using 8 frames.
|Poisson Boltzmann calculations performed using internal PBSA solver in
sander.
|
|All units are reported in kcal/mole.
|All entropy results have units kcal/mole (Temperature has been multiplied
in as 300 K).
DELTA S total= -24.2806 +/- 8.8063
DELTA G binding = -13.7627 +/- 3.8614 0.3861
DELTA G binding = -13.7627 +/- 3.8614 0.3861
-------------------------------------------------------------------------------
-------------------------------------------------------------------------------
Using Normal Mode Entropy Approximation: DELTA G binding = 10.5179 +/-
9.6157
-- Thank you very much I am looking forward to hearing from you Best wishes, Nguyen Ngoc Hong Phuc Email: phucnguyen20072.gmail.com Mobile phone: +84 984620799
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: rmsd-dexan-1.png)
