Date: Thu, 15 Sep 2011 21:56:35 -0700
Hi Francois,
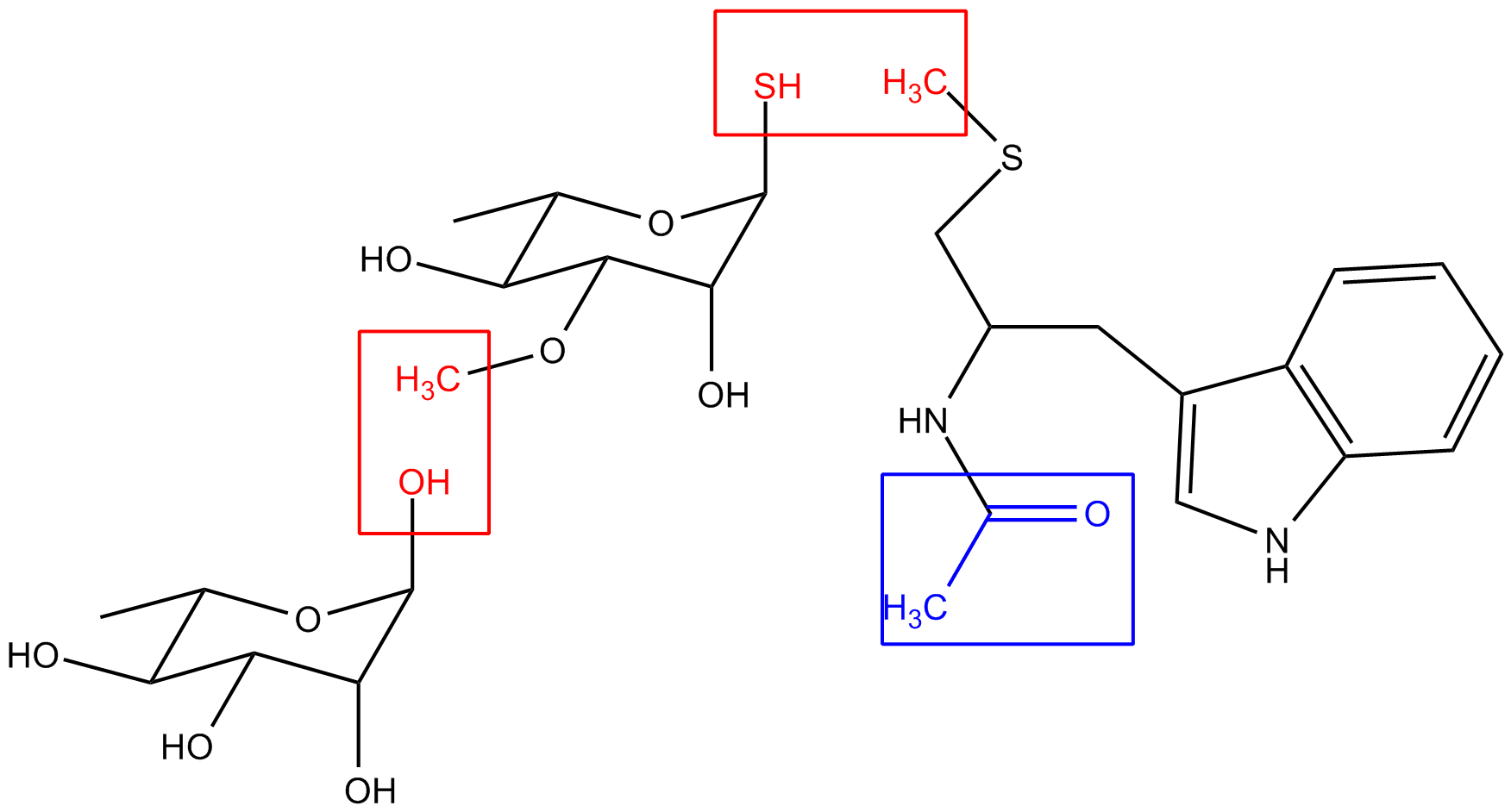
1.
Please see attached my charge-derivation scheme for one of the thiol-linked
'glycopeptides', where I have two red intermolecular charge constraints and
one blue intra-molecular charge constraint set to 0.
I will have 2 conformations for each sugar molecule respectively, and maybe
3 for the modified TRP. Does this look OK? I will follow the so-called
multi-molecule multi-conformation approach?
2.
Which .dat version of GLYCAM04 was used for developing q4md-CD and testing
in the cyclodextrin paper? And GLYCAM06 version?
3.
I tried deriving charges with R.E.D. server for one molecule. Without any
constraint, I found that the total charge of every single atom within this
molecule is 0.0004 e. Is this normal?
It seems the RRMS value can be used to see how close the derived charges fit
the ESP, but where should I find this value when running charge derivation
with R.E.D. server?
Thanks,
Yun
On Tue, Sep 13, 2011 at 8:46 AM, FyD <fyd.q4md-forcefieldtools.org> wrote:
> Yun,
>
> > And GLYCAM used different charges for alpha and beta anomers, so should I
> > stick to this even if I am using the 'amber' strategy?
>
> You do not 'need' to stick to this: you simply use two different
> building blocks: one is alpha and the other is beta; in these
> conditions, this is normal that by fitting to the MEP you get two
> different charge values for the alpha and beta anomers...
>
> regards, Francois
>
>
> > On Mon, Sep 12, 2011 at 11:32 PM, Yun Shi <yunshi09.gmail.com> wrote:
> >
> >> Hi Francois,
> >>
> >> I am worried since I saw no atom names information in the gaussian input
> >> file, which I used to do geometry optimization. I guess I should
> construct
> >> the gaussian input file from the xxx.pdb file generated by Ante_R.E.D.,
> then
> >> do my geometry optimization, then submit the gaussian out file together
> with
> >> xxx.p2n for charge derivation. So would there be any problem by doing
> so?
> >>
> >> In addition, I have only two major conformations for my small test
> molecule
> >> after QM geometry optimizations. So I wonder if I could just construct a
> >> merged p2n file with pdb coordinates from the two major QM-optimized
> >> conformers? I don't quite understand why R.E.D. server need QM
> optimization
> >> output file.
> >>
> >> Looking through the 'q4md-CD' paper, I feel it kind of counter-intuitive
> to
> >> mix GLYCAM04 geometrical parameters with atomic charges from 'Amber'
> >> strategy, which however, reproduced experimental data so well. But now I
> >> feel like that the RESP charges come hand-in-hand with the scaling
> factor,
> >> and I guess using the amber scaling factors is the major reason that
> your
> >> charge-derivation approach is successful.
> >>
> >> Since my molecule (the thio-glycosidic part) has no reference to look
> for
> >> experimental data to compare, what would be a simple experiment to carry
> out
> >> to validate my parameters? Or just try fitting them to QM calculations?
> >>
> >> By the way, what do you mean by " 'recompute' key dihedrals" when taking
> >> parameters from GAFF? And which are key dihedrals?
> >>
> >> Thanks,
> >>
> >> Yun
> >>
> >>
> >>
> >> On Wed, Sep 7, 2011 at 11:57 PM, FyD <fyd.q4md-forcefieldtools.org>
> wrote:
> >>
> >>> Dear Yun,
> >>>
> >>> > I basically understand this approach now. But for this
> >>> Rha-S-CH2-CH(R)-NH-
> >>> > group, how should I deal with the N terminus? If I simply add an ACE
> cap
> >>> to
> >>> > make it Rha-S-CH2-CH(R)-NH-ACE,
> >>>
> >>> Yes, with a trans peptide bond:
> >>>
> >>> H
> >>> Rha-S-CH2-CH(R)-N-C-Me
> >>> O
> >>>
> >>> > then how to set charge constraints?
> >>>
> >>> You set a single INTRA-MCC = 0 for the ACE chemical group in the P2N
> file.
> >>>
> >>> See http://q4md-forcefieldtools.org/Tutorial/Tutorial-3.php#15
> >>> (However in this #15 example two capping groups are added, so two
> >>> INTRA-MCC are set in the P2N file; In your case, you need to set a
> >>> single INTRA-MCC in your P2N file).
> >>>
> >>> > Should I
> >>> > add a Met or a Hydrogen to O3 on Rha? Because CENTRAL fragments
> instead
> >>> of
> >>> > TERMINNAL fragments are needed here.
> >>>
> >>> So you need two L-Rhamnoside building blocks...
> >>> a perOH-thiomethyl-Rhamnoside & a perOH-O-methyl-Rhamnoside
> >>>
> >>> [You could also try a different set of connecting groups between
> >>> S/O-hemiacetals (instead of S/O-acetals) and methoxy groups (i.e.
> >>> ether groups instead of hydroxyl groups).
> >>>
> >>> > Sorry our library does not have access to the Cieplak et al. paper.
> >>>
> >>> ok, I will send you the pdf file to your personal address (I think the
> >>> license to publish exclude the free distribution of this paper)...
> >>>
> >>> > In addition, I noted that GLYCAM06 have exactly the same charges for
> >>> most
> >>> > atoms in the same sugar molecule with different linkage, i.e.,
> >>> > 2-L-alpha-Rhamnose share identical atomic charges with
> >>> 3-L-alpha-Rhamnose
> >>> > except O2 and O3. However in amber99sb, every corresponding single
> atom
> >>> of,
> >>> > say TRP, would have different charges from ntTRP. I guess this may
> have
> >>> > something to do with the charge-deriving methods, as qwt=0.0005/0.001
> in
> >>> > 99sb VS qwt=0.01 in GLYCAM.
> >>>
> >>> This has to do with the charge constraints applied during the charge
> >>> fitting step.
> >>>
> >>> > But then the question would be, should I set charge constraints to
> most
> >>> > atoms on the sugar ring except the anomeric carbon and derive charges
> >>> using
> >>> > RESP-C2 for the sugar part, which would allow me to avoid the use of
> the
> >>> > first Rhamnose? Or just do all charges with RESP-A1A?
> >>>
> >>> Well you are right; this is the key ;-)
> >>>
> >>> -1- I guess GLYCAM developers will suggest you to use something
> >>> similar to the RESP-C2 charge model; i.e. MEP computation using the
> >>> CHELPG algo. and a single RESP fitting stage using the qwt=.01
> >>> hyperbolic restraint. This means using 1.0 scaling factors for the 1-4
> >>> non-bonding interactions for the sugar part of your ligand and 1.2/2.0
> >>> for the other part. Quite complex... considering the size of your
> >>> ligand and considering that the Connolly surface algorithm is used in
> >>> MEP computation and a two RESP stage fitting approach is used for the
> >>> non-sugar part.
> >>>
> >>> -2- Considering that L-Rhamnose is the 6-deoxy-L-mannose, I would
> >>> follow the approach we described .
> >>> http://www.ncbi.nlm.nih.gov/pubmed/21792425/ for all your ligand and
> >>> more general all your molecular system (in case of a Galacto
> >>> configuration I would suggest you to use the GLYCAM approach -1-).
> >>> This means 1.2/2.0 scaling factors for all the 1-4 non-bonding
> >>> interactions and using the Connolly surface algorithm in MEP
> >>> computation and a two RESP stage fitting approach for the entire
> >>> ligand. Far more simple... However, this approach has obviously to be
> >>> validated (however as it is requested for the case -1-).
> >>>
> >>> regards, Francois
> >>>
> >>>
> >>>
> >>> > On Sat, Aug 27, 2011 at 12:16 AM, FyD <fyd.q4md-forcefieldtools.org>
> >>> wrote:
> >>> >
> >>> >> Dear Yun,
> >>> >>
> >>> >> - When you have a peptide bond within a molecule, R1NH-COR2 you
> could
> >>> >> split this molecule into two parts R1NH-ACE & NME-COR2 (ACE = CH3CO;
> >>> >> NME = NHCH3; are capping groups). You use two INTRA-MCC set to zero
> >>> >> for these capping groups:
> >>> >>
> >>> >> R.E.D.:
> >>> >> 2 FG2 fragments LEaP
> >>> >> R1NH-ACE + NME-COR2 --> R1NH COR2 -----> R1NH-COR2
> >>> >> <=> <=>
> >>> >> 2 INTRA-MCC = 0
> >>> >>
> >>> >>
> >>> >> - When you have a disaccharide R'-O-R", you could split it into two
> >>> >> monosaccharides by using an INTER-MCC set to zero between the methyl
> >>> >> group of the methylglycoside (building-block 1) and the chosen
> >>> >> hydroxyl group belonging to the second monosaccharide unit:
> >>> >>
> >>> >> R.E.D.:
> >>> >> R'O-Me HO-R" 2 FG1 fragments LEaP
> >>> >> <-----> ---> R'O R" -----> R'-O-R"
> >>> >> 1 INTERMCC = 0
> >>> >>
> >>> >> by analogy, you could test:
> >>> >>
> >>> >> R'S-Me HS-R"
> >>> >> <-----> ---------------> R'-S-R"
> >>> >> 1 INTERMCC = 0
> >>> >>
> >>> >> With these simples rules you can split your red-blue-red-green
> >>> >> molecule into 4 building blocks.
> >>> >>
> >>> >> These have been defined some time ago; see
> >>> >> http://q4md-forcefieldtools.org/Tutorial/Tutorial-1.php
> >>> >> &
> >>> >> Cieplak et al. Application of the multimolecule and
> >>> >> multiconformational RESP methodology to biopolymers: Charge
> derivation
> >>> >> for DNA, RNA, and proteins. J. Comput. Chem. 1995, 16, 1357-1377.
> >>> >>
> >>> >> With R.E.D. Server/R.E.D. IV you can generate all these fragments
> into
> >>> >> a single R.E.D. job by using the corresponding P2N files and by
> >>> >> defining the correct constraints during the charge fitting step.
> >>> >> See http://q4md-forcefieldtools.org/Tutorial/Tutorial-3.php
> >>> >> & http://q4md-forcefieldtools.org/Tutorial/Tutorial-3.php#29
> >>> >>
> >>> >> regards, Francois
> >>> >>
> >>> >>
> >>> >> Quoting Yun Shi <yunshi09.gmail.com>:
> >>> >>
> >>> >> > Sorry that I should have attached this model molecule I want to
> study
> >>> >> > earlier.
> >>> >> >
> >>> >> > As you can see, I divide this ligand into four parts, and the
> >>> parameters
> >>> >> > (atomic charges, bonds, angles, dihedrals, impropers) of the red
> >>> >> (terminal)
> >>> >> > sugar and green ASP can be obtained from GLYCAM and 99SB
> >>> respectively.
> >>> >> >
> >>> >> > But I have to include the glycosidic oxygen between two sugar
> rings
> >>> in
> >>> >> the
> >>> >> > blue thio sugar, as this glysicidic oxygen is defined in the
> >>> preceding
> >>> >> sugar
> >>> >> > (the blue thio sugar) instead of the succeeding sugar according to
> >>> >> GLYCAM.
> >>> >> >
> >>> >> > I plan to use the approach -2- you mentioned (so that I don't need
> >>> >> generate
> >>> >> > many conformations), but considering my specific case, I wonder
> why
> >>> not
> >>> >> > using
> >>> >> >
> >>> >> > Me-O3-L-RhamnoS-Me + MeS-CH2CH(R)NH-COMe
> >>> >> > <----------------------> <===========>
> >>> >> > INTER-MCC=0 INTRA-MCC=0
> >>> >> >
> >>> >> > , and then remove the Me - Me and MeS - COMe respectively?
> >>> >> >
> >>> >> > Since the L-Rha and TRP in my case carry only one modified site
> >>> compared
> >>> >> to
> >>> >> > their standard counterparts, should I include charge constraints
> >>> (keep
> >>> >> the
> >>> >> > atomic charges) for atoms far away from the modified sites? Such
> as
> >>> C5
> >>> >> and
> >>> >> > C6 in the thio sugar, and the entire indole ring of the modified
> TRP?
> >>> >> >
> >>> >> > Thank you so much!
> >>> >> >
> >>> >> > Yun
> >>> >> >
> >>> >> >
> >>> >> > On Thu, Aug 25, 2011 at 11:37 PM, FyD <
> fyd.q4md-forcefieldtools.org>
> >>> >> wrote:
> >>> >> >
> >>> >> >> Yun,
> >>> >> >>
> >>> >> >> > Sorry, I just read the tutorial, that is, "Central fragment of
> a
> >>> xxx".
> >>> >> >> >
> >>> >> >> > So for the modified (C-terminal reduced) amino acid, should I
> add
> >>> a
> >>> >> MeCO
> >>> >> >> at
> >>> >> >> > the N-terminus and MeS at the reduced C-terminus? (considering
> the
> >>> >> >> reduced
> >>> >> >> > C-terminus is used to link glycosidic sulfur atom).
> >>> >> >> >
> >>> >> >> > And for the sugar molecule, should I add a Me cap to 3-hydroxyl
> >>> group
> >>> >> in
> >>> >> >> > addition to a methyl group attached to the glycosidic sulfur?
> >>> (This
> >>> >> sugar
> >>> >> >> > molecule would fall into the category of central fragment of my
> >>> >> ligand)
> >>> >> >>
> >>> >> >> ups I overlooked your problem ;-) You do not have any cysteine
> >>> >> residue...
> >>> >> >>
> >>> >> >> Let's re-do the explanations with your terminal molecule/pseudo
> >>> >> amino-acid:
> >>> >> >>
> >>> >> >> -1- you consider the following molecule
> >>> >> >>
> >>> >> >> Thio-L-Rhamno-CH2CH(R)NH-COMe ; R = CH2-Ph
> >>> >> >> <==>
> >>> >> >> INTRA-MCC
> >>> >> >>
> >>> >> >> You only have to set up an intra-molecular charge constraint
> >>> >> >> (INTRA-MCC keyword) for the COMe capping group (see <==> above).
> >>> >> >> See http://q4md-forcefieldtools.org/Tutorial/Tutorial-3.php#15
> >>> >> >>
> >>> >> >> R.E.D. will generate a sm fragment (mol2 file) for you.
> >>> >> >>
> >>> >> >> -2- you split your molecule into two building blocks.
> >>> >> >>
> >>> >> >> L-RhamnoS-Me + HS-CH2CH(R)NH-COMe
> >>> >> >> <-------> <==>
> >>> >> >> INTER-MCC INTRA-MCC
> >>> >> >>
> >>> >> >> See http://q4md-forcefieldtools.org/Tutorial/Tutorial-3.php#16
> >>> >> >> or http://q4md-forcefieldtools.org/Tutorial/Tutorial-3.php#17
> >>> >> >>
> >>> >> >> You set up an intra-molecular charge constraint (INTRA-MCC
> keyword)
> >>> >> >> for the COMe capping group.
> >>> >> >> You set up an inter-molecular charge constraint (INTER-MCC
> keyword)
> >>> >> >> between the methyl group of L-RhamnoS-Me and the thiol group of
> your
> >>> >> >> pseudo terminal amino-acid.
> >>> >> >>
> >>> >> >> The approach -2- is once again more complex but more flexible...
> >>> >> >>
> >>> >> >> R.E.D. will generate a FG fragment (mol2 file) in the Mol_MM
> >>> directory.
> >>> >> >>
> >>> >> >> Sorry for misunderstanding your problem in my first email.
> >>> >> >>
> >>> >> >> regards, Francois
> >>> >> >>
> >>> >> >>
> >>> >> >> > On Thu, Aug 25, 2011 at 9:58 AM, Yun Shi <yunshi09.gmail.com>
> >>> wrote:
> >>> >> >> >
> >>> >> >> >> Thank you very much!
> >>> >> >> >>
> >>> >> >> >> I looked at the CD project, and I saw when FFTopDB were
> >>> constructed,
> >>> >> the
> >>> >> >> >> 1-metylated glucose was used to derive RESP charges. So in my
> >>> >> >> >> thio-glycopeptide case, should I use the sugar molecule with a
> >>> methyl
> >>> >> >> group
> >>> >> >> >> attached to the glycosidic sulfur as well?
> >>> >> >> >>
> >>> >> >> >> What about the modified (C-terminal reduced) amino acid? Add
> one
> >>> >> more
> >>> >> >> >> methyl group to the modified C-terminal and acetate to the
> >>> >> N-terminal?
> >>> >> >> >>
> >>> >> >> >> But in the end, what is deposited in the FFTopDB is the
> residue
> >>> >> without
> >>> >> >> the
> >>> >> >> >> methyl or acetate cap, right?
> >>> >> >> >>
> >>> >> >> >> Yun
> >>> >> >> >>
> >>> >> >> >>
> >>> >> >> >>
> >>> >> >> >> On Wed, Aug 24, 2011 at 11:38 PM, FyD <
> >>> fyd.q4md-forcefieldtools.org
> >>> >> >> >wrote:
> >>> >> >> >>
> >>> >> >> >>> Dear Yun Shi,
> >>> >> >> >>>
> >>> >> >> >>> > I am trying to understand how this works.
> >>> >> >> >>>
> >>> >> >> >>> If you look at the data available in the "F-85" R.E.DD.B.
> >>> project,
> >>> >> you
> >>> >> >> >>> will find a x/tLEaP script to construct the CD-based
> >>> glycopeptides
> >>> >> as
> >>> >> >> >>> well as a frcmod file for missing force field parameters with
> >>> >> comments.
> >>> >> >> >>>
> http://q4md-forcefieldtools.org/REDDB/projects/F-85/script1.ff
> >>> >> >> >>>
> http://q4md-forcefieldtools.org/REDDB/projects/F-85/script3.ff
> >>> >> >> >>>
> >>> >> >> >>> > So instead of combining individual residues in a
> >>> building-block
> >>> >> >> manner,
> >>> >> >> >>> as
> >>> >> >> >>> > in the assignment of atomic charges for proteins with
> >>> amber99sb,
> >>> >> it
> >>> >> >> is
> >>> >> >> >>> > recommended to consider the ligand as a holistic molecule
> when
> >>> >> >> >>> calculating
> >>> >> >> >>> > the RESP charge?
> >>> >> >> >>>
> >>> >> >> >>> 'recommended'? ;-) ... Personally, I use most of the time the
> >>> >> building
> >>> >> >> >>> block approach whatever if the target 'big' molecule is a
> ligand
> >>> or
> >>> >> a
> >>> >> >> >>> nucleic acid/protein/polysacharide.
> >>> >> >> >>>
> >>> >> >> >>> > I am curious that if I could do things in a building-block
> >>> >> >> >>> > manner since it can potentially decrease a lot of
> >>> computational
> >>> >> time
> >>> >> >> for
> >>> >> >> >>> > geometry optimization.
> >>> >> >> >>>
> >>> >> >> >>> The building-block approach has many advantages:
> >>> >> >> >>> - it potentially "decreases a lot of computational time for
> >>> geometry
> >>> >> >> >>> optimization" as you said.
> >>> >> >> >>> - it allows rigorously defining the conformation of each
> >>> >> >> >>> building-block and not to use a conformation more or less
> >>> randomly
> >>> >> >> >>> chosen.
> >>> >> >> >>> - it allows avoiding interactions between charges group
> during
> >>> >> >> >>> geometry optimization in gas phase.
> >>> >> >> >>> - it allows the construction of analogs for the target
> molecule.
> >>> >> >> >>> - it allows the construction of oligomers/polymers for the
> >>> target
> >>> >> >> >>> molecule.
> >>> >> >> >>>
> >>> >> >> >>> However, it also has disadvantages:
> >>> >> >> >>> - it is complex to set up when one starts, but R.E.D. has
> been
> >>> >> >> >>> designed for this approach.
> >>> >> >> >>> - errors during the charge fitting step are introduced when
> >>> using
> >>> >> the
> >>> >> >> >>> building-block approach; these errors have to be minimized by
> >>> >> >> >>> correctly selecting the connecting groups between the
> different
> >>> >> >> >>> building-blocks. The statistics module available in R.E.D.
> >>> >> >> >>> Server/R.E.D. IV also helps to localize/minimize these
> errors.
> >>> >> >> >>>
> >>> >> >> >>> > And when it comes to geometrical parameters, we should use
> >>> GLYCAM
> >>> >> for
> >>> >> >> >>> sugar
> >>> >> >> >>> > part, 99SB for standard amino acids, and GAFF for organic
> >>> part?
> >>> >> >> >>>
> >>> >> >> >>> Yes
> >>> >> >> >>>
> >>> >> >> >>> - We only select 'obvious' missing force field parameters
> from
> >>> GAFF
> >>> >> >> >>> (we recompute key dihedrals), and when used we always
> >>> rationalize
> >>> >> >> >>> these force field parameters as it was done in the Cornell at
> >>> al.
> >>> >> >> >>> force field.
> >>> >> >> >>>
> >>> >> >> >>> - In this work, we used Amber scaling factor values for 1-4
> >>> >> >> >>> non-bonding interactions for all the glycopeptide molecular
> >>> systems;
> >>> >> >> >>> i.e. we did not split the system into a peptide and a sugar
> >>> parts.
> >>> >> >> >>>
> >>> >> >> >>> > BTW, could you tell me how to generate multiple
> conformations
> >>> with
> >>> >> >> >>> geometry
> >>> >> >> >>> > optimization from Gaussian 09?
> >>> >> >> >>>
> >>> >> >> >>> You could do a conformational search - although if the
> >>> >> building-block
> >>> >> >> >>> approach is used the conformational search is quite
> >>> simplified...
> >>> >> >> >>> We also often modify a key dihedral to look for lowest
> >>> >> minimum/minima.
> >>> >> >> >>>
> >>> >> >> >>> To create a P2N file with multiple conformations, see:
> >>> >> >> >>> http://q4md-forcefieldtools.org/Tutorial/Tutorial-1.php#3
> >>> >> >> >>>
> >>> >> >>
> >>> >>
> >>>
> http://q4md-forcefieldtools.org/Tutorial/Tutorial-1.php#EXAMPLE-P2N-FILE
> >>> >> >> >>>
> >>> >> >> >>> To create a QM file with multiple conformations to be used in
> >>> the
> >>> >> Mode
> >>> >> >> >>> 2 of R.E.D. (see
> >>> >> >> >>> http://q4md-forcefieldtools.org/REDS/popup/popredmodes.php),
> >>> simply
> >>> >> >> >>> concatenate the different QM outputs into a single file.
> >>> >> >> >>>
> >>> >> >> >>> regards, Francois
> >>> >> >> >>>
> >>> >> >> >>> >> Dear Yun,
> >>> >> >> >>> >>
> >>> >> >> >>> >> > Is it technically possible to do it due diligence in the
> >>> first
> >>> >> >> place?
> >>> >> >> >>> >> That
> >>> >> >> >>> >> > is, cut the molecule into three parts as I mentioned
> >>> before,
> >>> >> use
> >>> >> >> >>> GLYCAM
> >>> >> >> >>> >> for
> >>> >> >> >>> >> > the sugar part, 99SB for the Thr, and GAFF for modified
> Phe
> >>> and
> >>> >> >> the
> >>> >> >> >>> >> > thio-glycosidic linkage. And may I then link these parts
> >>> >> together
> >>> >> >> >>> using
> >>> >> >> >>> >> LEaP
> >>> >> >> >>> >> > ?
> >>> >> >> >>> >>
> >>> >> >> >>> >> Concerning the use of GLYCAM + GAFF + Amber99SB you might
> be
> >>> >> >> >>> >> interested by looking at the following paper:
> >>> >> >> >>> >> http://www.ncbi.nlm.nih.gov/pubmed/21792425
> >>> >> >> >>> >> & its corresponding R.E.DD.B. project .
> >>> >> >> >>> >> http://q4md-forcefieldtools.org/REDDB/projects/F-85/ +
> its
> >>> LEaP
> >>> >> >> >>> script:
> >>> >> >> >>> >>
> >>> http://q4md-forcefieldtools.org/REDDB/projects/F-85/script1.ff
> >>> >> >> >>> >>
> >>> >> >> >>> >> This work is about cyclodextrin based-glycopeptide and 1-4
> >>> >> >> non-bonding
> >>> >> >> >>> >> interactions in GLYCAM & Amber99SB.
> >>> >> >> >>> >>
> >>> >> >> >>> >> Your structure is not a cyclodextrin but this work
> describe
> >>> (i)
> >>> >> how
> >>> >> >> to
> >>> >> >> >>> >> derive charges and build force field libraries for new
> >>> fragments
> >>> >> by
> >>> >> >> >>> >> using R.E.D. IV and (ii) proposes new directions
> concerning
> >>> the
> >>> >> >> >>> >> treatment of 1-4 non-bonding interactions in the context
> of
> >>> >> >> >>> >> glycopeptides.
> >>> >> >> >>> >>
> >>> >> >> >>> >> Finally, in the LEaP script you will find examples how to
> >>> connect
> >>> >> >> >>> >> organic, amino-acid and monosaccharide units...
> >>> >> >> >>> >>
> >>> >> >> >>> >> regards, Francois
>
>
>
>
> _______________________________________________
> AMBER mailing list
> AMBER.ambermd.org
> http://lists.ambermd.org/mailman/listinfo/amber
>
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: ChargeConstraint.png)
