Date: Tue, 28 Sep 2010 11:18:25 +0530
Hi All,
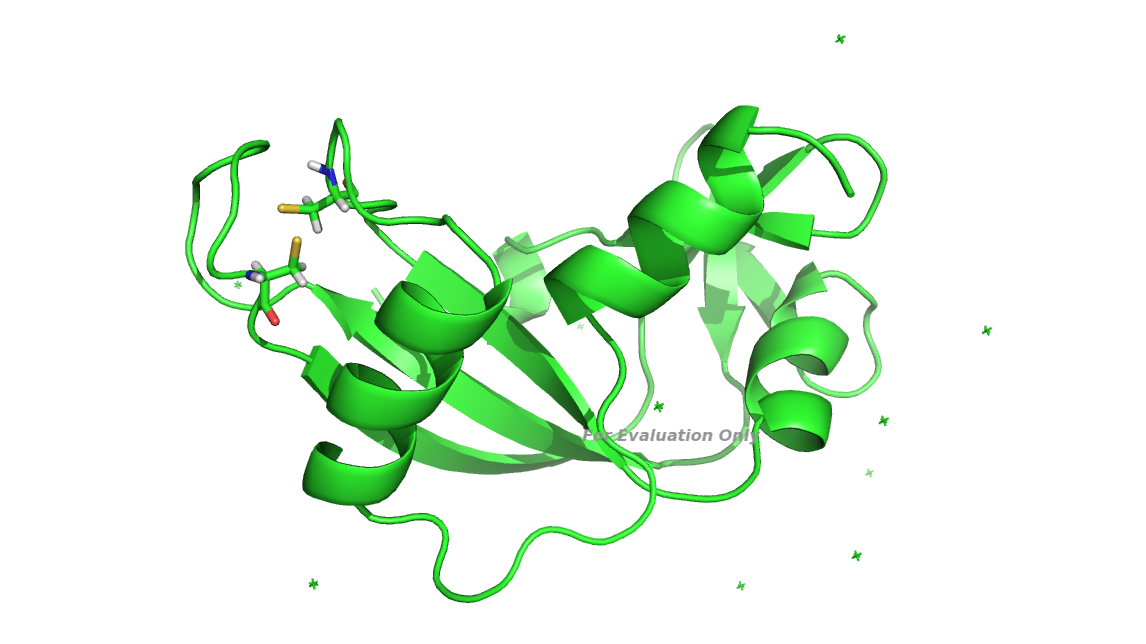
In my protein of interest, there is a disulfide bond in between two residues
so I prepared the .pdb structure by converting *CYS to CYX* and then I used
the* bond command* in tleap for the intact cystine formation. I generated
the pdb structure after tleap module preparation, there was cystine in the
structure. But Once I was done with minimization, the cystine was again
converted to two cysteine (disulfide bond is breakage). I want to keep the
disulphide bond intact during minimization and equilibration. I am wondering
is there any way to keep the disulphide bond intact during minimization and
equilibration.
Below mentioned is the input file for the minimization. I have also attached
the image of the broken cystine.
Minimization with Cartesian restraints for the solute
&cntrl
imin=1, maxcyc=1000, ncyc=500,
ntpr=100,
cut =10.0,
ntr=1,
&end
Group input for restrained atoms
100.0
RES 1 122
END
END
Hirdesh
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: 111.png)
