Date: Fri, 27 Feb 2009 19:21:29 +0100
Dear amber users,
I would like to know if there is any possibility how to calculate entropy
change (complexate X noncomplexate state) in the case
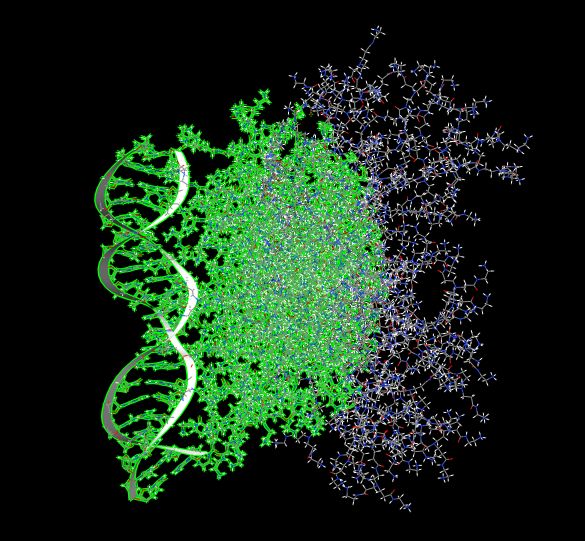
of relatively big complexes (cca 15k atoms), like for example high
generation dendrimer + oligonucleotide - please see the attached picture.
I didn't succeed with direct calculation of the whole complex entropy due
to the enormous RAM memory requirements (36 GB).
At the end I tried it with NAB on the PC with 32 GB RAM but my calculation
was interrupt after 4 days without finishing the entropy calculation.
(only minimisation was done).
So my question is: is it possible to calculate entropy of the whole
complex using some "Per-Partes" methodology or is it
possible to at least calculate (as an sufficient approximation) just the
entropy of some relevant specific region (in my attached picture the green
part).
If this is not available in Amber ( using MM-PBSA, NAB ) does anybody know
about some another software where is possible to realize such calculations
?
Thanks in advance for any relevant information/directions.
Marek
-- Tato zpráva byla vytvořena převratným poštovním klientem Opery: http://www.opera.com/mail/
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/jpeg attachment: complex.jpg)
