Date: Mon, 24 Mar 2008 11:59:24 -0400
you're right, that doesn't seem to be working correctly.
can you send me directly your sander input? also do you have
anything else in the system except the 1 peptide and water?
On Mon, Mar 24, 2008 at 11:51 AM, Geoff Wood <geoffrey.wood.epfl.ch> wrote:
> Dear Amber Community,
> I am curious about the imaging done in hybrid remd calculations.
>
> If I use ptraj commands (see below) to post process a trajectory then the
> imaged trajectory looks like what one would expect (see attached picture).
> However, if I look at the stripped restart file then the imaging seems to
> be a bit strange (see the other attached picture). Could anyone comment on
> this?
>
>
> ptraj commands:
>
> *trajin coords*
> *center * mass origin*
> *image origin center*
> *solvent byname WAT TIP3*
> *closest 350 :mask first*
> *trajout coords.strip nobox*
>
> Thanks,
>
> Dr Geoffrey Wood
> Ecole Polytechnique Fédérale de Lausanne
> SB - ISIC - LCBC
> BCH 4108
> CH - 1015 Lausanne
>
>
>
>
-- =================================================================== Carlos L. Simmerling, Ph.D. Associate Professor Phone: (631) 632-1336 Center for Structural Biology Fax: (631) 632-1555 CMM Bldg, Room G80 Stony Brook University E-mail: carlos.simmerling.gmail.com Stony Brook, NY 11794-5115 Web: http://comp.chem.sunysb.edu ===================================================================
-----------------------------------------------------------------------
The AMBER Mail Reflector
To post, send mail to amber.scripps.edu
To unsubscribe, send "unsubscribe amber" to majordomo.scripps.edu
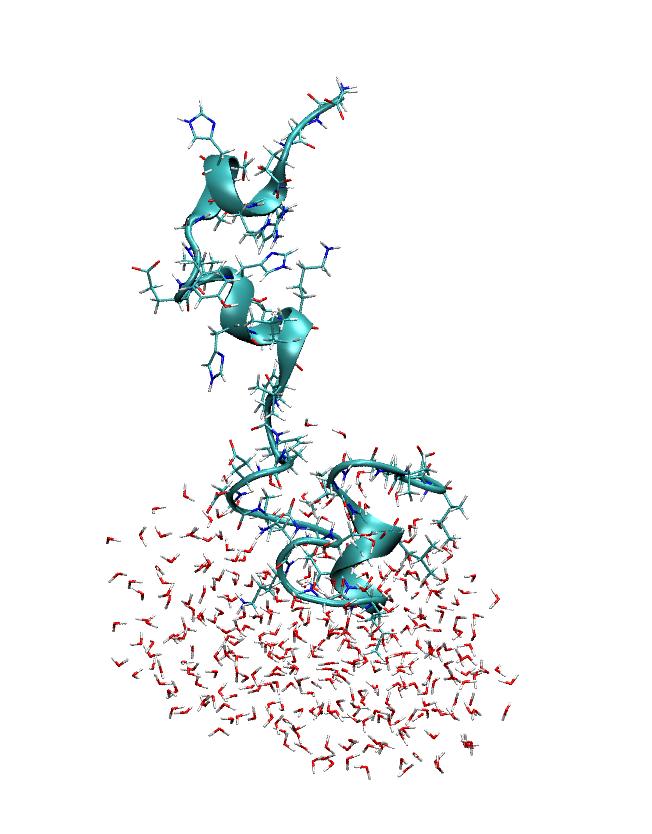
(image/jpeg attachment: sanderRestart.jpeg)
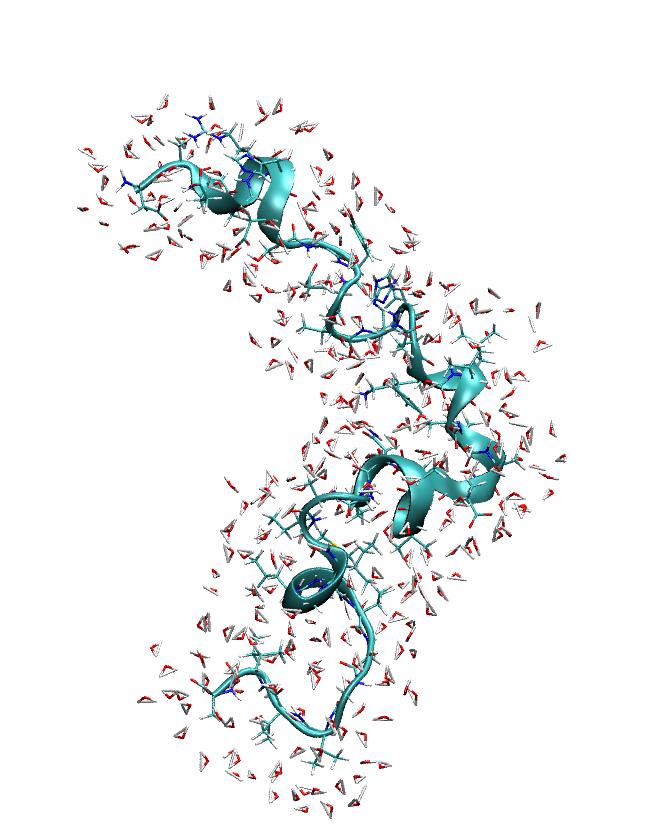
(image/jpeg attachment: ptrajout.jpeg)
