Date: Tue, 19 Jun 2007 08:47:58 -0400
Hello All!
>
> Following are the input files I used to study a system of protein
> surrounded by drugs (parameterized using antechamber).
>
> ==============
> First I minimized the system with restraints on residues 1-420 which were
> picked from a previous simulation which was stabilized over a few
> nanoseconds at 312 K. This part (1-420) was picked and surrounded with more
> water and drugs.
> ==============
> minmization
> &cntrl
> imin = 1,
> maxcyc = 5000,
> ntpr = 100,
> ncyc = 500,
> ntr = 1,
> restraint_wt = 2.0,
> restraintmask = ':1-420'
> /
> ================
> Then I heated the system from 250K to 400K again with the first 420
> residues restrained.
> ==============
> 500ps heating with NVT
> &cntrl
> imin = 0,
> irest = 0,
> nstlim = 500000,
> dt = 0.001,
> tempi = 250,
> temp0 = 400,
> cut = 8,
> ntb = 1,
> ntt = 1,
> ntf = 2,
> ntc = 2,
> ntpr = 1000,
> ntwr = 1000,
> ntwx = 1000,
> ntwe = 1000,
> ntr = 1,
> restraint_wt = 2.0,
> restraintmask =':1-420'
> /
> =========================
> Cooling the system to 312K with restraints on first 420 residues
> =========================
> 100ps cooling with NVT
> &cntrl
> imin = 0,
> irest = 1,
> ntx = 5,
> nstlim = 100000,
> dt = 0.001,
> tempi = 402.37,
> temp0 = 312,
> cut = 8,
> ntb = 1,
> ntt = 1,
> ntf = 2,
> ntc = 2,
> ntpr = 1000,
> ntwr = 1000,
> ntwx = 1000,
> ntwe = 1000,
> ntr = 1,
> restraint_wt = 2.0,
> restraintmask =':1-420'
> /
> ==========================
> NPT at 312K on the whole system. No restraints on any part of the system
> ==========================
> 200ps NPT
> &cntrl
> imin = 0,
> irest = 1,
> ntx = 5,
> nstlim = 200000,
> dt = 0.001,
> tempi = 312,
> temp0 = 312,
> cut = 8,
> ntb = 2,
> ntp = 1,
> taup = 2.0,
> ntt = 1,
> ntf = 2,
> ntc = 2,
> tautp = 1,
> ntpr = 1000,
> ntwr = 1000,
> ntwx = 1000,
> ntwe = 1000,
> /
> ==========================
>
> All the plots, temperature, Etot, etc. don't have any major fluctuations.
> The Ewald error estimate for NPT had a mean of 5*10^(-5) and a standard
> deviation of 3*10(-5). Should this be a cause for alarm?
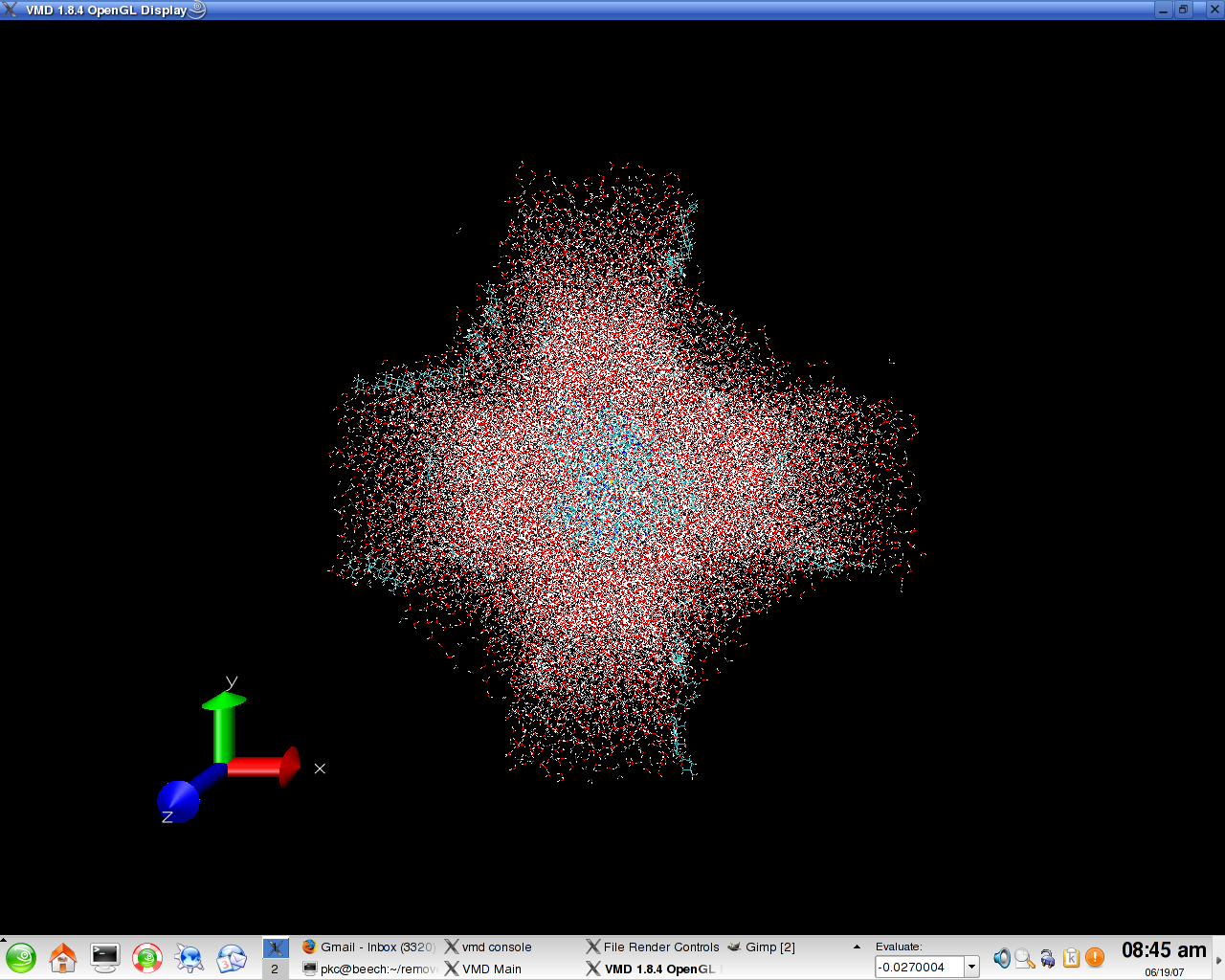
> When I imaged the frames to center the initial 420 residues I ended up
> with a system which was fine when looked at two planes (rectangular).
> However when viewed it along the third plane, it looked like a plus symbol.
> (Figure attached).
> Could you please comment on why it turned out that way. I don't see a
> reason why it should have the edges filletted and even if should, why only
> in one direction?
> Please let me know if you want any additional information.
>
> Thank you,
> Pavan K. Ghatty
>
>
-----------------------------------------------------------------------
The AMBER Mail Reflector
To post, send mail to amber.scripps.edu
To unsubscribe, send "unsubscribe amber" to majordomo.scripps.edu
(image/png attachment: snapshot1.png)
