Date: Thu, 27 Mar 2025 12:04:55 +0530 (IST)
Dear AMBER Community,
I am performing umbrella sampling following the tutorial available at [ https://github.com/callumjd/AMBER-Umbrella_COM_restraint_tutorial | this link ] . My objective is to determine the free energy profile for the transfer of an ethanol molecule through a DMPC membrane bilayer. My system is similar to the tutorial, except that I have replaced methanol with ethanol while keeping the number of DMPC molecules and the box dimensions unchanged.
I followed all the steps as outlined in the tutorial. I generated 32 windows with 1 Å spacing and ran each window for 5 ns. After running WHAM, I observed that the PMF values suddenly increased after the 7th window, reaching unexpectedly high values up to the 32nd window. The histogram distribution appears satisfactory, but after applying diffusion correction, I obtained a PMF with negative values. I have attached all the plot.
I am unsure what mistake I might have made. Could anyone suggest how to resolve this issue?
Thank you in advance.
Best regards,
Sankar Maity
Research Scholar
NIT Rourkela
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
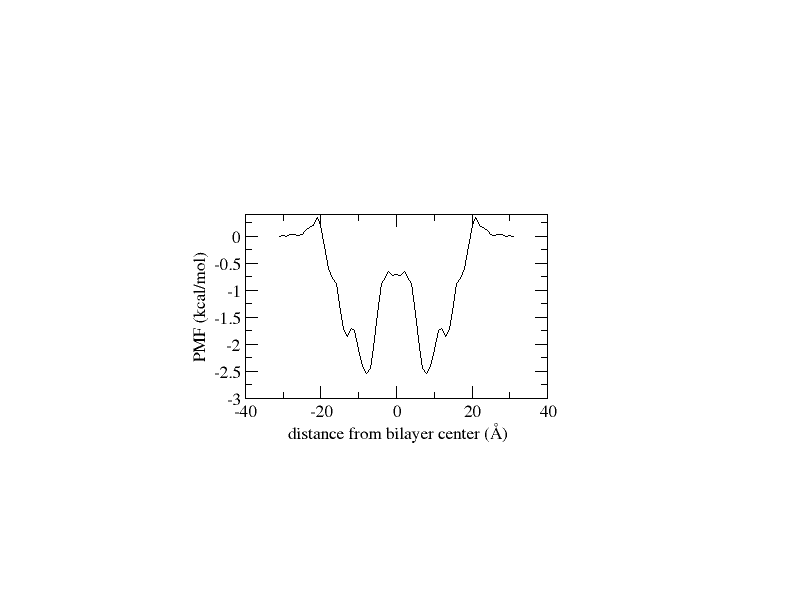
(image/png attachment: free_energy.png)
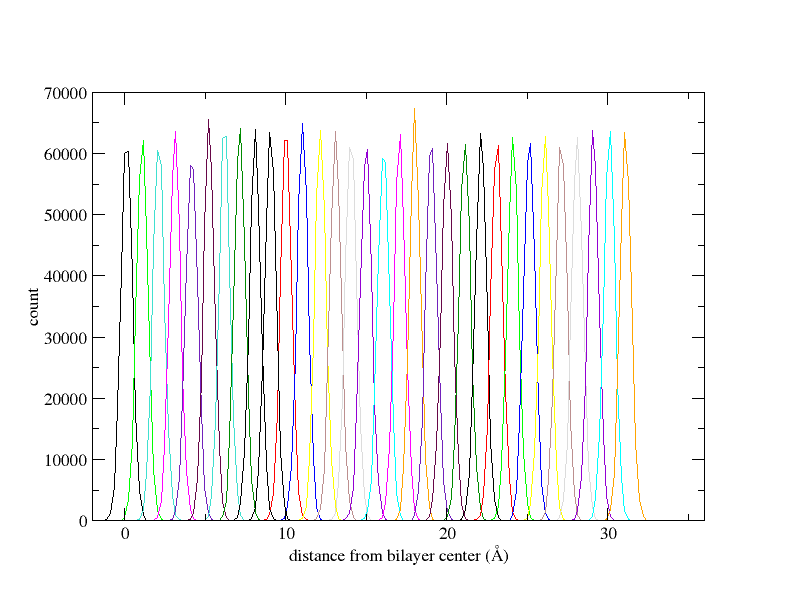
(image/png attachment: hist_plot.png)
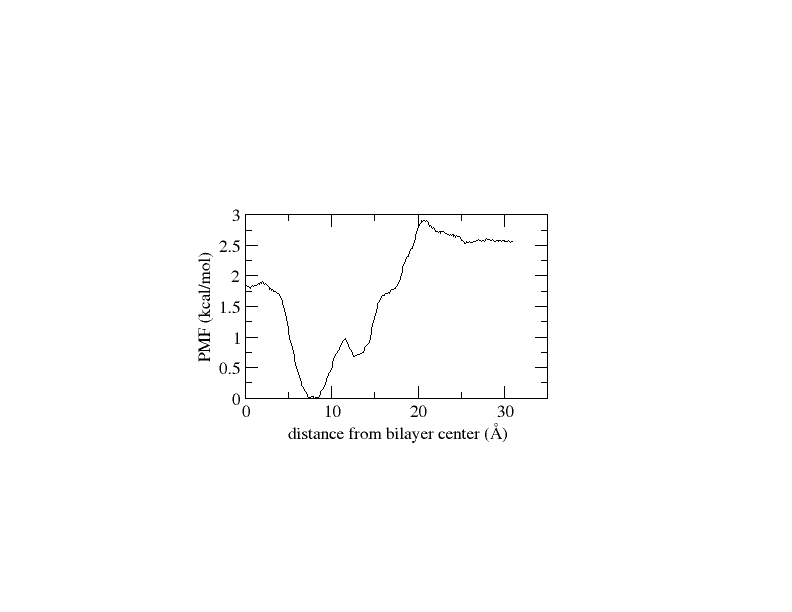
(image/png attachment: plot_free_energy.png)
