Date: Sun, 26 Jan 2025 16:21:36 +0000
Hi all
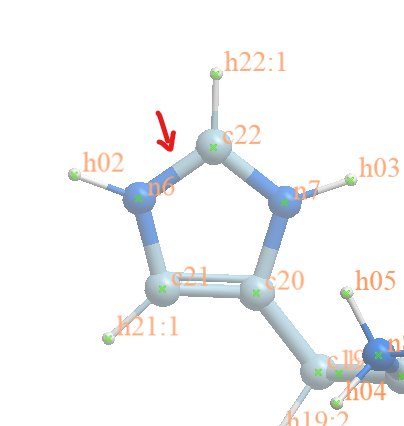
Part of my molecule has an imidazole and it is protonated.
When I take the docked pose (left picture in yellow carbons) and run ANTECHAMBER it seems to complain about N6 and N7 connectivity:
If I do not heed those warnings I get a output (see cyan picture) where I see the aromaticity is lost and two N+ atoms which is not what was the input.
Welcome to antechamber 22.0: molecular input file processor.
Info: acdoctor mode is on: check and diagnose problems in the input file.
Info: The atom type is set to gaff2; the options available to the -at flag are
gaff, gaff2, amber, bcc, and sybyl.
-- Check Format for mol2 File --
Status: pass
Warning: Ignoring Mol2 record type (.<TRIPOS>CRYSIN).
-- Check Unusual Elements --
Status: pass
-- Check Open Valences --
Warning: The number of bonds (3) for atom (ID: 68, Name: N6) does not match
the connectivity (2) for atom type (N.ar) defined in CORR_NAME_TYPE.DAT.
Warning: The number of bonds (3) for atom (ID: 72, Name: N7) does not match
the connectivity (2) for atom type (N.ar) defined in CORR_NAME_TYPE.DAT.
But, you may safely ignore the warnings if your molecule
uses atom names or element names as atom types.
-- Check Geometry --
for those bonded
for those not bonded
Status: pass
-- Check Weird Bonds --
Status: pass
-- Check Number of Units --
Status: pass
acdoctor mode has completed checking the input file.
[cid:1d77c6c9-e455-4c67-9851-05fc6eb6cd84][cid:7ddd6e64-501a-435b-af41-b74952312e95]
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber

(image/png attachment: image.png)
(image/png attachment: 02-image.png)
