Date: Thu, 26 Sep 2024 10:58:11 +0800
Dear all,
I am trying to perform minimization for one photosystem embedding with POPG
lipid bilayer membrane. First 900 cycles of CPU minimization using pmemd
and the next 60000 cycles of GPU minimization using pmemd.cuda can work
well. But when switched to the last 50000 cycles of CPU parallel
minimization using pmemd.MPI, this task failed.
The associated files we used are attached. Please take a look at this issue
here. Thanks!
01_Min.in: 400 cycles of CPU minimization; OK
02_Min.in: 500 cycles of CPU minimization; OK
03_Min.in: 10000 cycles of GPU minimization: OK
04_Min.in: 50000 cycles of GPU minimization: OK
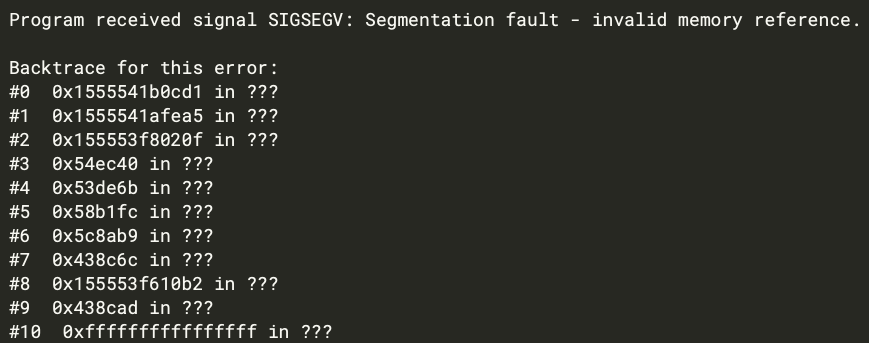
05_Min.in: 50000 cycles of CPU parallel minimization: FAILED, erro info
like:
[image: image.png]
mailinglist.tar.gz
<https://drive.google.com/file/d/1pH9LgVPVNmyNWohu_AmV0jDfx9H2Tzhy/view?usp=drive_web>
Best wishes,
Pengfei Li
----------------------------------------
Senior Application Scientist at
Single Particle, LLC
pengfei.li.singleparticle.com
https://www.singleparticle.com <http://singleparticle.cn/>
https://twitter.com/singleparticle
Single Particle, your one-stop shop for structural biology needs
[image: image.png]
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: image.png)
