From: Yongxian Wu via AMBER <amber.ambermd.org>
Date: Sat, 11 May 2024 19:47:19 -0700
Hi Mayukh,
I tested Amber 24 with Libtorch 2.0.1 and CUDA 11.7, and I could not
reproduce the error you encountered. In my tests, both pbsa_bcopt and
pbsa_saopt passed without any segmentation errors. Here is a screenshot of
my testing results.
[image: 1.png]
However, I did find a memory issue with pbsa, although it seems unrelated
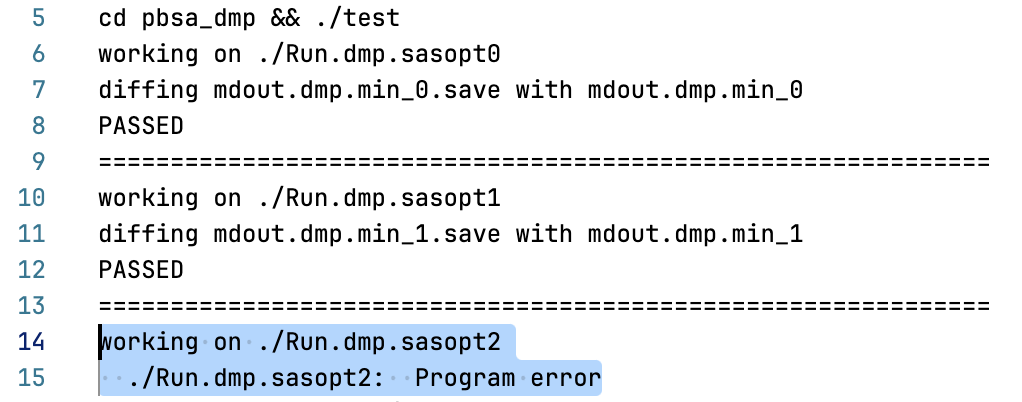
to Libtorch. When I run multiple pbsa testing processes simultaneously, the
pbsa_dmp test case fails due to an invalid memory reference.
[image: 2.png]
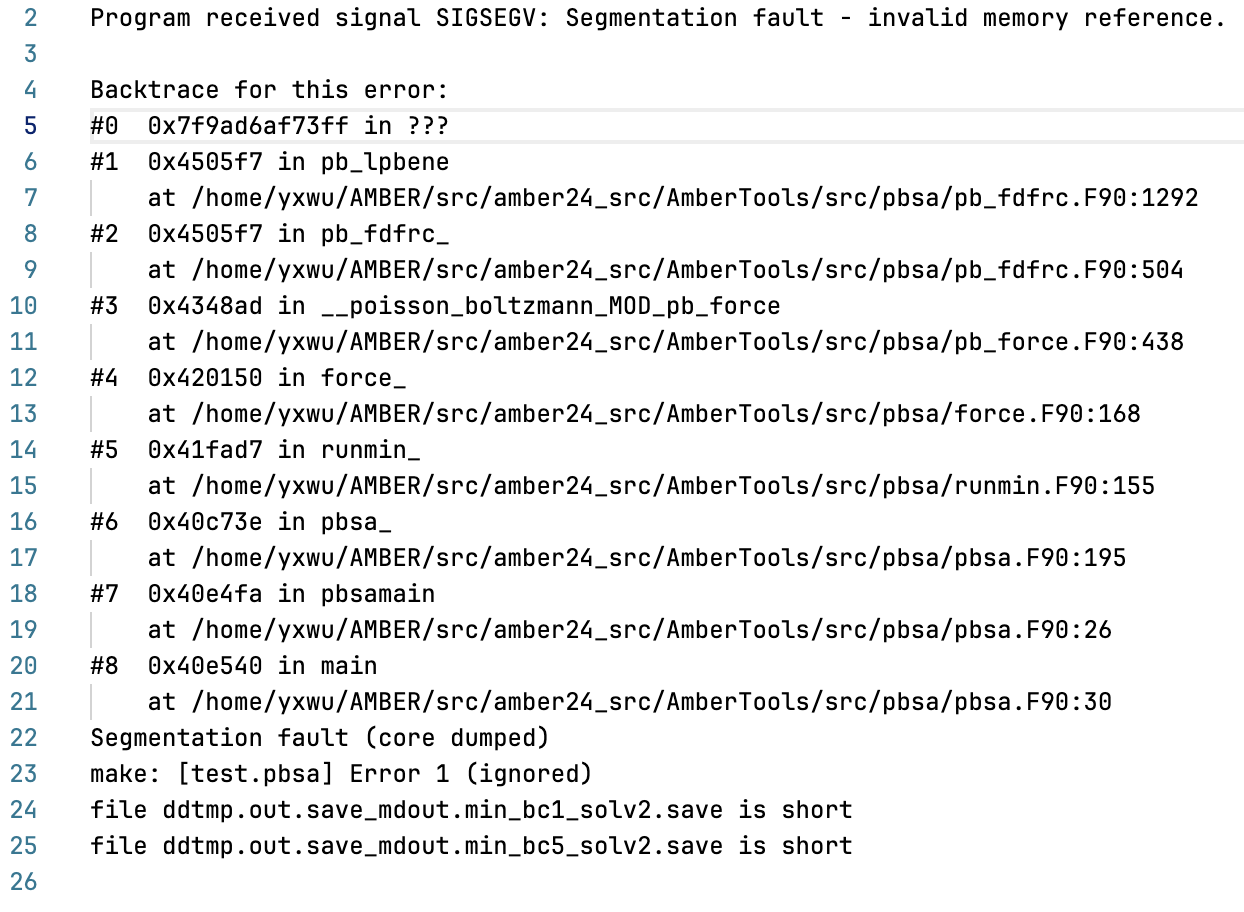
I attempted to compile Amber with debugging symbols to obtain readable
backtrace information. The error message appears as follows:
[image: 3.png]
Based on my initial investigation, I believe this problem is caused by a
Fortran memory issue and occurs only when multiple tests are run
simultaneously; otherwise, this test case would pass smoothly without any
errors.
To solve your problem, I suggest the following steps:
1. Please align your environment settings with mine to avoid any
unexpected errors. You can try using Libtorch 2.0.1 + CUDA 11.7, as I have
already tested it. If you have CUDA 11.8 installed, it should be quite
straightforward to switch to CUDA 11.7.
2. Since the backtrace information attached in your email is unreadable,
you might consider recompiling Amber 24 with debugging symbols to get a
readable error message. To do this, simply add -DCMAKE_BUILD_TYPE=Debug to
your cmake command.
Let me know if you need further assistance.
Best,
Yongxian Wu
On Fri, May 10, 2024 at 7:07 AM Chakrabarti, Mayukh (NIH/NCI) [C] <
mayukh.chakrabarti.nih.gov> wrote:
> Hi Yongxian,
>
>
>
> Thank you for testing this. As mentioned in my earlier message, I
> re-tested by compiling a version of Amber24 with Pytorch 1.12.1 and CUDA
> 10.2. I used ‘libtorch-shared-with-deps-1.12.1+cu102.zip’, which
> corresponds to the specific CUDA version. As you mentioned, the compilation
> and build process proceeds without any issues. However, while the ‘make
> test.serial’ runs and reports the final testing statistics when it
> completes, it still reports errors when testing the pbsa binaries, e.g.:
>
>
>
> *cd pbsa_bcopt && ./Run.dmp.min*
>
>
>
> *Program received signal SIGSEGV: Segmentation fault - invalid memory
> reference.*
>
>
>
> *Backtrace for this error:*
>
> *#0 0x7ff0f7ca0171 in ???*
>
> *#1 0x7ff0f7c9f313 in ???*
>
> *#2 0x7ff0f735cc0f in ???*
>
> *#3 0x7ff15479a219 in ???*
>
> *#4 0x7ff0f735f856 in ???*
>
> *#5 0x7ff15477b722 in ???*
>
> *Segmentation fault (core dumped)*
>
> * ./Run.dmp.min: Program error*
>
> *make[2]: [Makefile:199: test.pbsa] Error 1 (ignored)*
>
>
>
> *cd pbsa_saopt && ./Run.dmp.min*
>
>
>
> *Program received signal SIGSEGV: Segmentation fault - invalid memory
> reference.*
>
>
>
> *Backtrace for this error:*
>
> *#0 0x7ff58f59f171 in ???*
>
> *#1 0x7ff58f59e313 in ???*
>
> *#2 0x7ff58ec5bc0f in ???*
>
> *#3 0x7ff5ec099219 in ???*
>
> *#4 0x7ff58ec5e856 in ???*
>
> *#5 0x7ff5ec07a722 in ???*
>
> *Segmentation fault (core dumped)*
>
> * ./Run.dmp.min: Program error*
>
>
>
> I ran ‘ldd’ on both the *pbsa* and *pbsa.cuda *binaries, and there don’t
> appear to be any missing linked libraries. You mentioned that “*Libtorch
> should still function even if you encounter errors with pbsa_test*”. Does
> this mean it is safe to ignore these SIGSEGV errors, assuming that libtorch
> is otherwise properly installed and corresponds to the CUDA version being
> used? Is it some issue with the test itself?
>
>
>
> Best,
>
>
>
> *Mayukh Chakrabarti *(he/him)
>
> COMPUTATIONAL SCIENTIST
>
>
>
> *From: *Yongxian Wu <yongxian.wu.uci.edu>
> *Date: *Friday, May 10, 2024 at 1:56 AM
> *To: *Chakrabarti, Mayukh (NIH/NCI) [C] <mayukh.chakrabarti.nih.gov>,
> AMBER Mailing List <amber.ambermd.org>
> *Cc: *Ray Luo <rluo.uci.edu>
> *Subject: *Re: [AMBER] [EXTERNAL] Re: Amber22/AmberTools23: Enabling of
> libtorch & cudnn libraries breaks pbsa binaries
>
> Hi Mayukh,
>
>
>
> I have tested compiling Amber24 with CUDA 11.7.0 and PyTorch 2.0.1. The
> compilation and build process proceeded without any issues, and make
> test.serial passed without any stopping errors. Please ensure you use the
> version of libtorch that corresponds to the specific CUDA version. However,
> note that the pbsa_test did not involve libtorch or MLSES. Libtorch should
> still function even if you encounter errors with pbsa_test. Here are the
> specific variables I used:
>
>
> -DLIBTORCH=ON \
>
> -DCUDA_HOME=$CUDA_PATH/cuda_11.7.0 \
>
> -DTORCH_HOME=$LIBTORCH_PATH/libtorch \
>
> -DCUDA_TOOLKIT_ROOT_DIR=/$CUDA_PATH/cuda_11.7.0 \
>
> -DCUDNN=TRUE \
>
> -DCUDNN_INCLUDE_PATH=$CUDNN_PATH/include \
>
> -DCUDNN_LIBRARY_PATH=$CUDNN_PATH/lib/libcudnn.so \
>
>
>
>
>
> Best regards,
>
> Yongxian Wu
>
>
>
> On Wed, May 8, 2024 at 2:26 PM Chakrabarti, Mayukh (NIH/NCI) [C] via
> AMBER <amber.ambermd.org> wrote:
>
> Hi Ray and Yongxian,
>
> Thank you for your response. I don’t have CUDA 11.3 available on my
> system, but I do have CUDA 11.8, which I tried to use instead. Upon your
> advice, I tried to re-compile Amber 24 & AmberTools 24 using an updated
> version of libtorch, version 2.1.0 with CUDA 11.8. I also used
> “cudnn-linux-x86_64-8.7.0.84_cuda11-archive.tar.xz”. Unfortunately,
> following compilation, I still encounter the same segmentation fault errors
> (SIGSEGV) in the pbsa binaries. I will next try with PyTorch 1.12.1 and
> CUDA 10.2, and report back.
>
> Best,
>
> Mayukh Chakrabarti (he/him)
> COMPUTATIONAL SCIENTIST
>
> From: Ray Luo <rluo.uci.edu>
> Date: Wednesday, May 8, 2024 at 1:04 PM
> To: Chakrabarti, Mayukh (NIH/NCI) [C] <mayukh.chakrabarti.nih.gov>, AMBER
> Mailing List <amber.ambermd.org>
> Subject: [EXTERNAL] Re: [AMBER] Amber22/AmberTools23: Enabling of libtorch
> & cudnn libraries breaks pbsa binaries
> Hi Mayukh,
>
> The issue might be due to the version of PyTorch being used.
>
> I recommend using the same version of libtorch that we currently have,
> which is version 1.12.1 with CUDA 11.3. PyTorch 1.9 with CUDA 10.2 is
> outdated. However, PyTorch 1.12.1 also supports CUDA 10.2, although we
> recommend using CUDA 11 since CUDA 10 is outdated.
>
> Let me know if there are any other questions.
>
> Best,
> Yongxian and Ray
> --
> Ray Luo, Ph.D.
> Professor of Structural Biology/Biochemistry/Biophysics,
> Chemical and Materials Physics, Chemical and Biomolecular Engineering,
> Biomedical Engineering, and Materials Science and Engineering
> Department of Molecular Biology and Biochemistry
> University of California, Irvine, CA 92697-3900
>
>
> On Tue, May 7, 2024 at 8:57 AM Chakrabarti, Mayukh (NIH/NCI) [C] via
> AMBER <amber.ambermd.org<mailto:amber.ambermd.org>> wrote:
> Hello,
>
> I wanted to update my report below to mention that the issue with pbsa
> binaries breaking upon enabling the LibTorch & cudnn libraries still
> persists in Amber24 with AmberTools 24. I compiled with CUDA 10.2, gcc 8.5,
> OpenMPI 4.1.5, python 3.10, and cmake 3.25.2 on a Red Hat Enterprise Linux
> release 8.8 (Ootpa) system. I am not aware of any workaround or resolution
> for this issue.
>
> Best,
>
> Mayukh Chakrabarti (he/him)
> COMPUTATIONAL SCIENTIST
>
> From: Chakrabarti, Mayukh (NIH/NCI) [C] <mayukh.chakrabarti.nih.gov
> <mailto:mayukh.chakrabarti.nih.gov>>
> Date: Tuesday, October 31, 2023 at 11:28 AM
> To: amber.ambermd.org<mailto:amber.ambermd.org> <amber.ambermd.org<mailto:
> amber.ambermd.org>>
> Subject: Amber22/AmberTools23: Enabling of libtorch & cudnn libraries
> breaks pbsa binaries
> Hello,
>
> I am encountering a problem in which enabling LibTorch libraries with
> Amber22 and AmberTools23 causes segmentation fault errors (SIGSEGV) in the
> pbsa binaries upon running the serial tests (make test.serial). I have
> successfully compiled a version of Amber22/AmberTools23 in which this
> library is not enabled, and none of the pbsa tests in AmberTools break
> (i.e., no segmentation fault errors).
>
> Further details:
>
> I am running my compilation with CUDA 10.2, gcc 8.5, OpenMPI 4.1.5, python
> 3.6, and cmake 3.25.1 on a Red Hat Enterprise Linux release 8.8 (Ootpa)
> system. As per the manual, I have tried both “Built-in” mode and
> “User-installed” mode, both resulting in the same errors. For the
> “User-installed” mode, I manually downloaded and extracted
> “libtorch-shared-with-deps-1.9.1+cu102.zip” from the PyTorch website to
> correspond to CUDA 10.2, and
> “cudnn-linux-x86_64-8.7.0.84_cuda10-archive.tar.xz” directly from the
> NVIDIA website, and specified the following variables to CMAKE:
>
>
> -DLIBTORCH=ON \
>
> -DTORCH_HOME=/path_to_libtorch \
>
> -DCUDNN=TRUE \
>
> -DCAFFE2_USE_CUDNN=1 \
>
> -DCUDNN_INCLUDE_PATH=/path_to_cudnn_include \
>
> -DCUDNN_LIBRARY_PATH=/path_to_libcudnn.so \
>
>
> Building and compiling proceed without issue. However, when running the
> serial tests, I get errors akin to the following (example shown after
> sourcing amber.sh and running AmberTools pbsa_ligand test):
>
> Program received signal SIGSEGV: Segmentation fault - invalid memory
> reference.
>
> Backtrace for this error:
> #0 0x7f480cb65171 in ???
> #1 0x7f480cb64313 in ???
> #2 0x7f480c221c0f in ???
> #3 0x7f4869667219 in ???
> #4 0x7f480c224856 in ???
> #5 0x7f4869648722 in ???
> ./Run.t4bnz.min: line 34: 2436022 Segmentation fault (core dumped)
> $DO_PARALLEL $TESTpbsa -O -i min.in
> <https://urldefense.com/v3/__http://min.in/__;!!CzAuKJ42GuquVTTmVmPViYEvSg!MVPl6p4KbMJpbOqO568Ns0Kz5i7sbGNJ-naGNHZf9Ycly3ycDbDToZ4J3VrVyvCP1-1IQPwcf2tM6Z4nwjoll1JKRHaF82Q$>
> <
> https://urldefense.com/v3/__http://min.in/__;!!CzAuKJ42GuquVTTmVmPViYEvSg!NC0Jwxkjnbm_mXLPxsbm3fva29jev44srLmSv7hv0XZuaw716Egbv1itPhJek6uNYpgJ3GbSbeK6jyZvAA$
> > -o $output < /dev/null
> ./Run.t4bnz.min: Program error
>
> When running the exact same test with the version of Amber22 not
> containing the LibTorch libraries:
>
> diffing mdout.lig.min.save with mdout.lig.min
> PASSED
> ==============================================================
>
> I have run ‘ldd’ on the pbsa binary to try to identify any libraries that
> may be missing, but there don’t appear to be any missing libraries. Could
> anybody please provide any insight into how to fix this issue?
>
> Best,
>
> Mayukh Chakrabarti
> COMPUTATIONAL SCIENTIST
>
>
>
>
>
>
>
> _______________________________________________
> AMBER mailing list
> AMBER.ambermd.org<mailto:AMBER.ambermd.org>
>
> https://urldefense.com/v3/__http://lists.ambermd.org/mailman/listinfo/amber__;!!CzAuKJ42GuquVTTmVmPViYEvSg!PLN2_ZzXkAP-XJnmppjnXaq9ruj3FNFdRR163QR8ehrjX6YYBOUfKc8Eo0t0w7N8I3CtOwXatyYNzg$
> CAUTION: This email originated from outside of the organization. Do not
> click links or open attachments unless you recognize the sender and are
> confident the content is safe.
>
> _______________________________________________
> AMBER mailing list
> AMBER.ambermd.org
>
> https://urldefense.com/v3/__http://lists.ambermd.org/mailman/listinfo/amber__;!!CzAuKJ42GuquVTTmVmPViYEvSg!NC0Jwxkjnbm_mXLPxsbm3fva29jev44srLmSv7hv0XZuaw716Egbv1itPhJek6uNYpgJ3GbSbeI1bOw3SQ$
>
> CAUTION: This email originated from outside of the organization. Do not
> click links or open attachments unless you recognize the sender and are
> confident the content is safe.
>
>
>
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
Received on Sat May 11 2024 - 20:00:03 PDT