Date: Fri, 9 Feb 2024 19:09:31 +0000
Hello RNA experts and modelers, I am very new to the RNA forcefields and simulations and I am trying to set up some MD on a RNA structure which has 2 modified uracils in the sequence. I have loaded the leap ff files as instructed and I am trying to see if someone can help me in parametrizing this modified residue.
is there a place/database I can look into for modified lib/frcmod files for modified bases in RNA?
> source leaprc.gaff2
> source leaprc.RNA.OL3
> source leaprc.modrna08
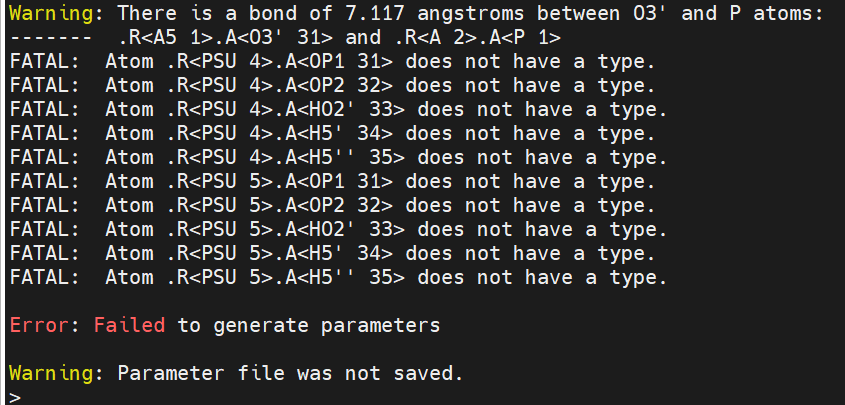
[cid:3cd96c5c-5b92-4ece-9181-55309560e2d5]
The pdb is 6HMO. If someone can give a bit of guidance it will be much appreciated.
Best,
Debarati
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: image.png)
