Date: Tue, 6 Feb 2024 23:42:38 +0000
Hello,
I have finished a constant pH simulation in explicit solvent.
One thing I noticed that is confusing to me:
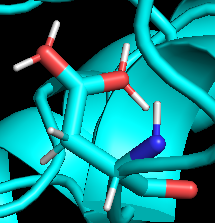
When I visualize the protein side chains, I found that the carboxylic acids on Asparate and Glutamate, have two hydrogens attaching to each of the oxygen. (as shown in the picture below: )
[cid:image001.png.01DA592B.5F7A0930]
I am confused about why this is happening. The hydrogens were added by tleap before I run the simulations.
I saw some comments here (http://archive.ambermd.org/201508/0476.html
) about 4-hydrogen carboxylate groups. But I donít understand why there are two hydrogens on a single oxygen.
Is this somehow supposed to be like this for AS4 and GL4 in constant pH?
Does the pdb file (converted from the trajectory files) from the constant pH reflect the actual protonation state?
Thank you very much!
Zoey
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: image001.png)
