Date: Wed, 24 Jan 2024 17:52:44 +0100
Dear Amber community !
I have another question related to post-treatment of MD trajectory
with membrane system.
This time dealing with the modeling of dimeric GPCR (fully embedded in
the membrane) I use the following script to remove PBC from the
trajectory.
parm *.prmtop
trajin *rod_*.nc 1 -1 10
trajout stripped_traj44.pdb
strip :WAT,Na+,Cl-
center :49,374 # center on the 2 residues involved in the prot-prot contact
autoimage
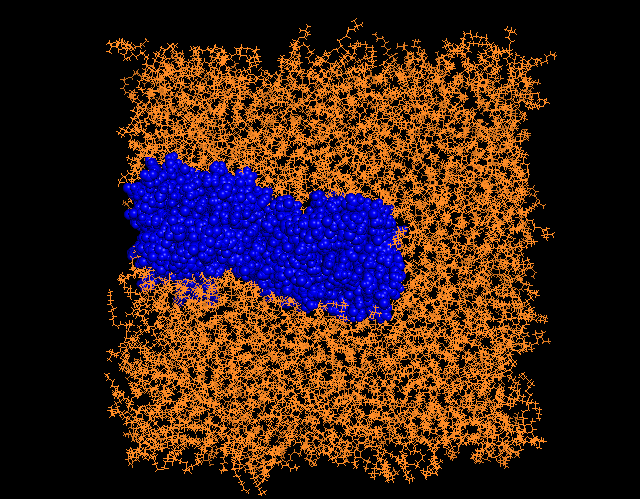
Briefly, it does everything correctly (no jumps across the pbs etc)
but the both proteins look a bit shifted (although I have tried to
center the view in my cpptraj script) towards one of the parts of the
membrane (please see the screenshot attached !). Would it be rather
possible to orient the protomers in order that it could be perfectly
in the center plane ?
Many thanks in advance !
Sincerely
Enrico
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: test_gpcr.png)
