Date: Tue, 6 Jun 2023 14:59:00 +0530
Dear amber users,
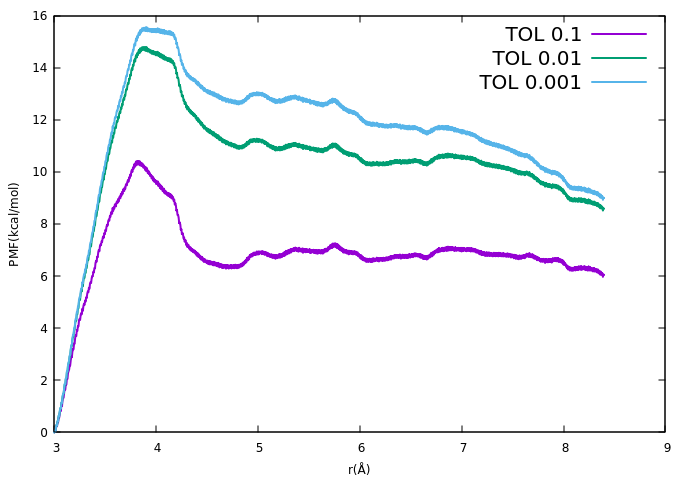
I am doing Umbrella sampling of an ion with a phosphate group of DNA.
While performing WHAM when I vary TOLERANCE value, my PMF values fluctuate
so much.
Following are those WHAM commands
>wham 3 8.4 1000 0.1 300 0 meta.dat result0.1.dat
>wham 3 8.4 1000 0.01 300 0 meta.dat result0.01.dat
>wham 3 8.4 1000 0.001 300 0 meta.dat result0.001.dat
The PMF profiles for these corresponding TOLERANCE values is the attached
graph.
Please find the attachment. I am not able to understand why on varying the
values of TOLERANCE the pmf profiles fluctuate this much.
*-------------------------------------------------*
*With Regards*
*Angad Sharma*
*Research Scholar*
*School of Computational & Integrative Sciences*
*Jawaharlal Nehru University*
*New Delhi 110067, (INDIA)*
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: tolerance.png)
