Date: Mon, 5 Jun 2023 02:33:37 +0530
Dear amber users,
I am doing umbrella sampling of DNA with an ion. From DNA I have taken a
fixed site of the phosphate group and my ion is fixed at 10 angstrom away
from this position of DNA. Here the reaction coordinate is distance.
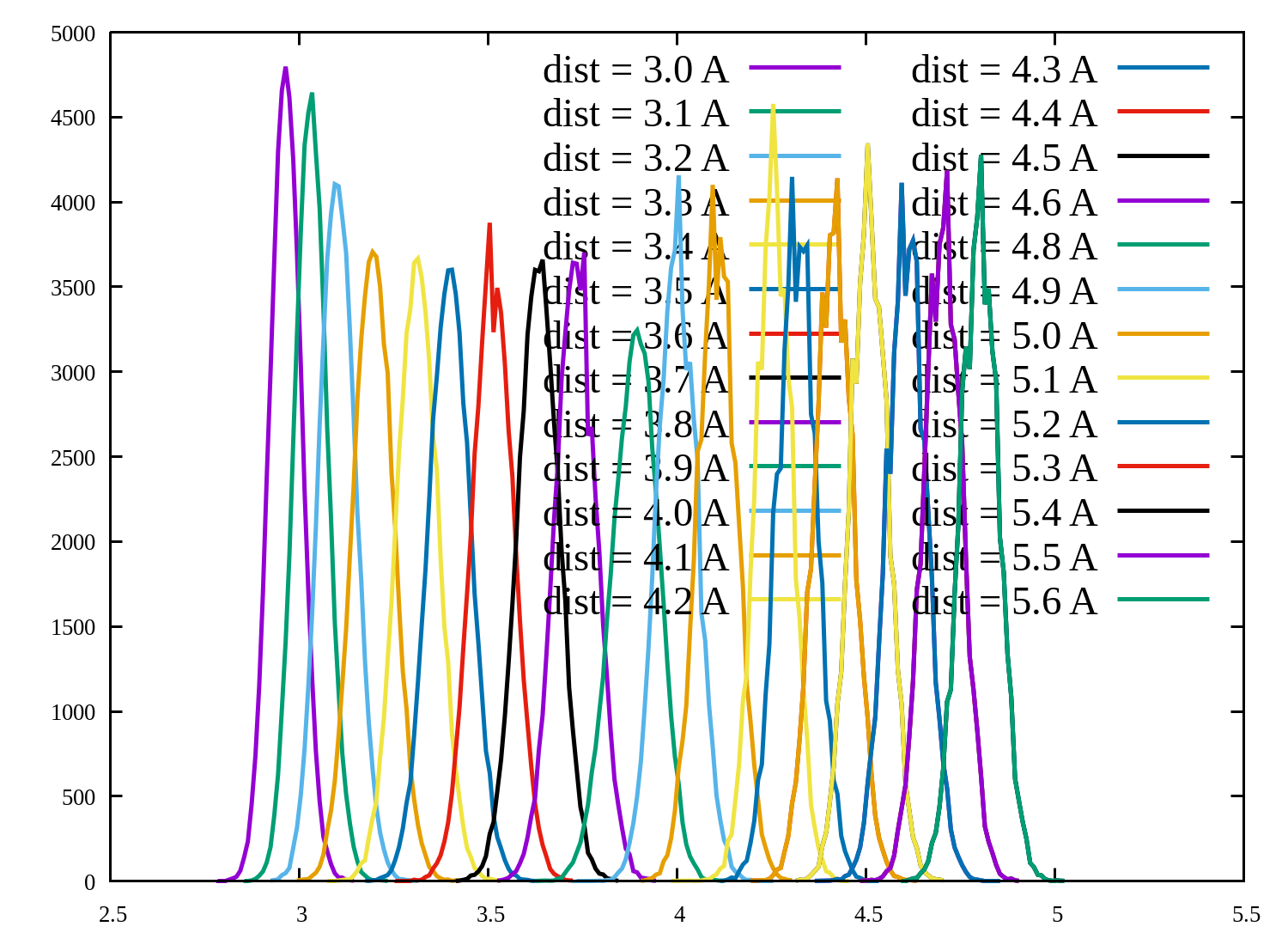
On plotting the histogram plot of windows it is coming fine. I am attaching
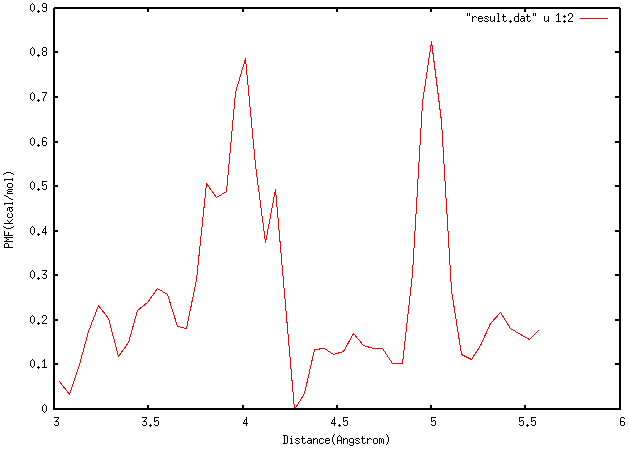
the plot, but the corresponding PMF is not coming correct. The WHAM command
for distance (reactive coordinate ) which i used for calculating PMF is as
the follows:-
> wham 3 7.5 46 0.001 300 0 meta.dat result.dat
I guess I am making a mistake in using the WHAM command.
Please help me to figure it out.
thanks
*------------------------------------------------*
*With Regards*
*Angad Sharma*
*Research Scholar*
*School of Computational & Integrative Sciences*
*Jawaharlal Nehru University*
*New Delhi 110067, (INDIA)*
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: histoi1.png)
(image/png attachment: pmf.png)
