Date: Sun, 22 Jan 2023 11:32:37 +0000
Dear Amber users,
I’m trying to run simulations for a 8-mer ssDNA with artificial nucleotides by GBn2 model. I ran 5000 steps of energy minimization and then equilibration. I encountered phosphate aggregation in the equilibration step, as shown in the attached image. I checked the terms in the output file and found the rms fluctuation of EELEC and EGB were very large, EELEC significantly increased and EGB significantly decreased. I’m wondering if someone ever met a similar problem and how to deal with it. For normal ssDNA(without artificial nucleotides) everything works well, it only happens when adding new nucleotides.
My prmtop/prmcrd files, min.in, equ.in files are attached.
Best regards,
Jiaan
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
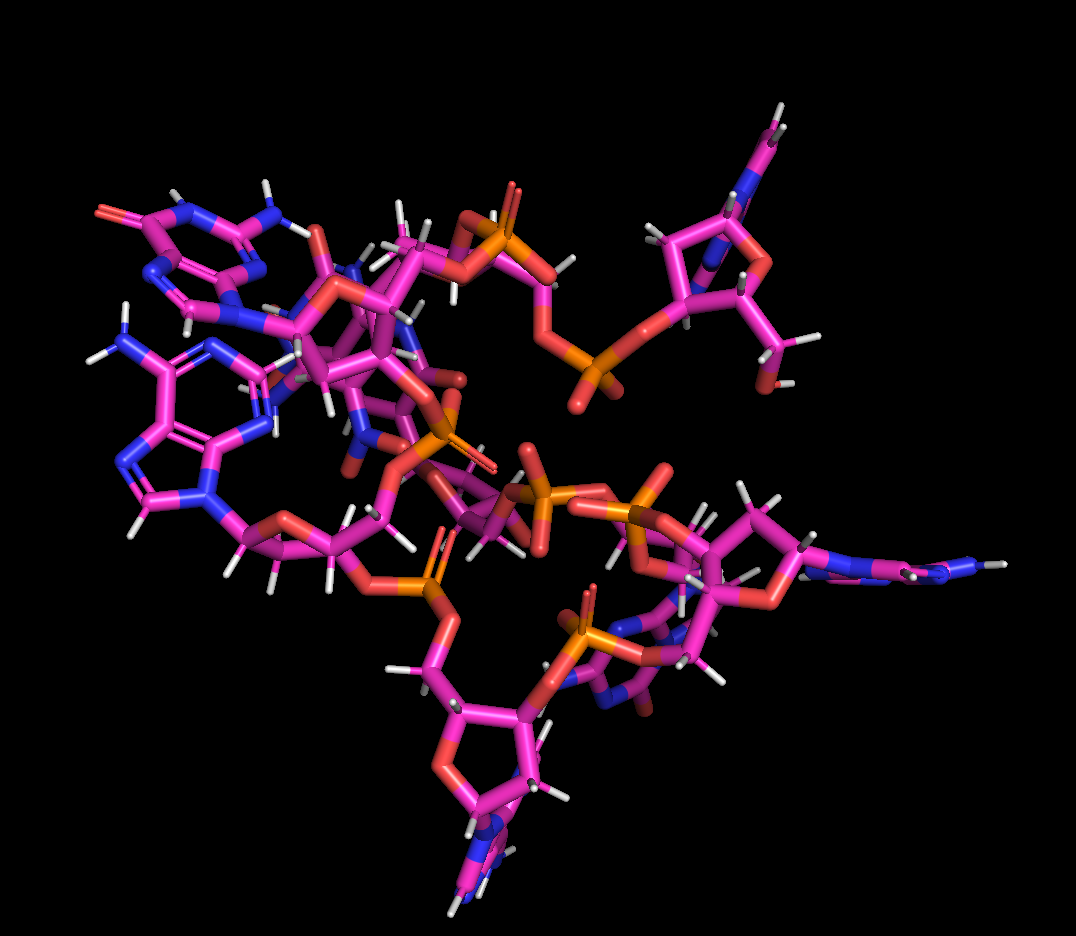
(image/png attachment: Phosphate_aggregation.png)
- application/octet-stream attachment: dna.prmtop
- application/octet-stream attachment: dna.rst7
- application/octet-stream attachment: min.in
- application/octet-stream attachment: equ.in
