Date: Tue, 17 Jan 2023 03:48:35 +0000
Hi,
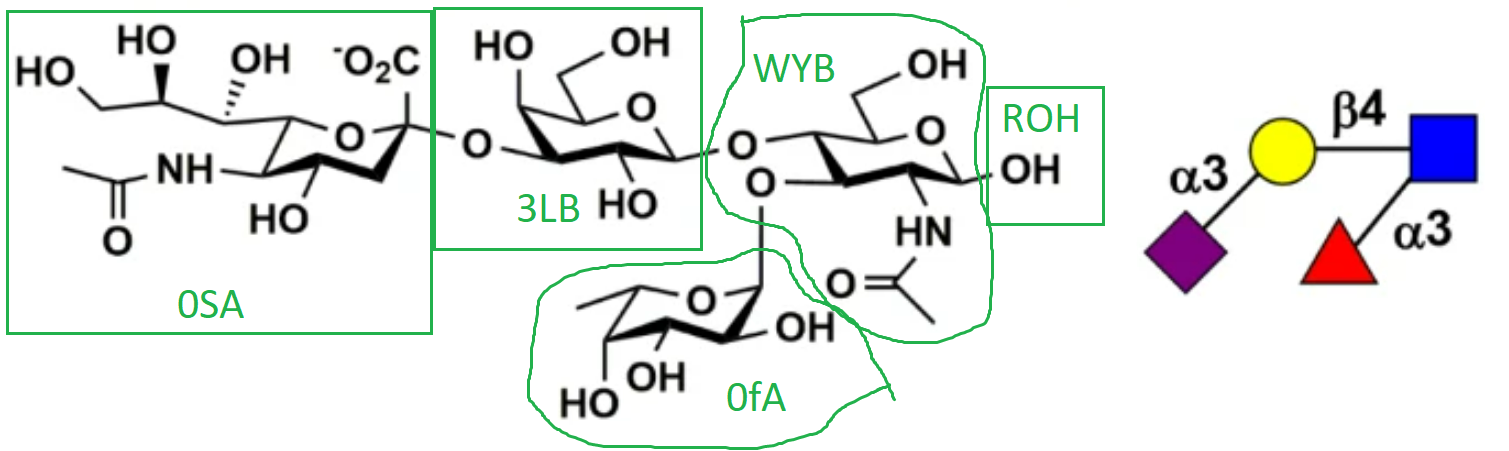
Recently I'm trying to simulate an oligosaccharide. The structure is shown in the figure and the corresponding GLYCAM residue name is also labeled (also attached).
[cid:image001.png.01D929F3.DEE59580]
I prepared the structure and renamed all the atoms and residues in accordance with the GLYCAM residues similar to what I usually do for amino acid residues.
When I use tleap to generate the topology files:
source leaprc.GLYCAM_06j-1
a = loadpdb Lig2.pdb
An error showed:
/dors/csb/apps/amber19/bin/teLeap: Error!
Comparing atoms
.R<0fA 3>.A<C2 18>,
.R<0fA 3>.A<H1 2>,
.R<0fA 3>.A<O5 3>, and
.R<3LB 2>.A<O3 21>
to atoms
.R<0fA 3>.A<C2 18>,
.R<WYB 1>.A<C1 1>,
.R<0fA 3>.A<O5 3>, and
.R<3LB 2>.A<O3 21>
This error may be due to faulty Connection atoms.
!FATAL ERROR----------------------------------------
!FATAL: In file [chirality.c], line 142
!FATAL: Message: Atom named C1 from WYB did not match !
!
!ABORTING.
And tleap was terminated. I checked the structure multiple times, but I do not understand why WYB C1 was identified as 0fA H1. Could you please provide some suggestions? I attached the pdb file of the oligosaccharide.
Thanks a lot!
Best,
Yaoyukun Jiang
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: image001.png)
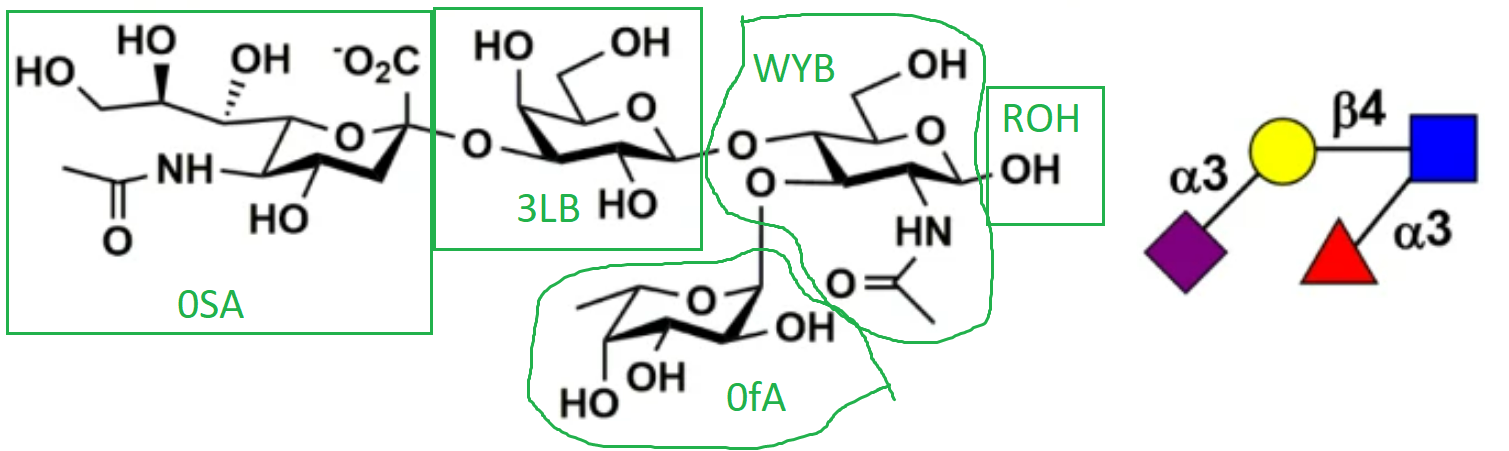
(image/png attachment: oligosaccharide.png)
- application/octet-stream attachment: Lig2.pdb
