Date: Tue, 8 Jun 2021 09:41:42 +0530
Hi All,
I'm trying to calculate the pair distribution function between the
residue 50 and water molecules within 2.4 angstrom from the residue 50.
I have used the following input:
*trajin ../product.nc <http://product.nc>*
*pairdist out pairdist.dat mask :50<.2.4:WAT*
I run cpptraj as
*cpptraj -p ../6pti_ions.prmtop < pairdist.in <http://pairdist.in> >
pairdist.out*
I got the following error in the terminal
*Error: No reference set, cannot select by distance.Error: No reference
set, cannot select by distance.*
However, I did not get any error message in the pairdist.out file
Below is the output of pairdist.out
CPPTRAJ: Trajectory Analysis. V16.00
___ ___ ___ ___
| \/ | \/ | \/ |
_|_/\_|_/\_|_/\_|_
| Date/time: 06/08/21 08:50:35
| Available memory: 245.129 MB
Reading '../6pti_ions.prmtop' as Amber Topology
INPUT: Reading input from 'STDIN'
[trajin ../product.nc]
Reading '../product.nc' as Amber NetCDF
[pairdist out pairdist.dat mask :50<.2.4:WAT]
PAIRDIST: Calculate P(r) for atoms selected by ':50<.2.4:WAT'
Output to 'pairdist.dat'
Resolution is 0.010000 Ang.
---------- RUN BEGIN -------------------------------------------------
PARAMETER FILES (1 total):
0: 6pti_ions.prmtop, 19924 atoms, 4846 res, box: Trunc. Oct., 4790 mol,
4751 solvent
INPUT TRAJECTORIES (1 total):
0: 'product.nc' is a NetCDF AMBER trajectory, Parm 6pti_ions.prmtop
(Trunc. Oct. box) (reading 50000 of 50000)
Coordinate processing will occur on 50000 frames.
BEGIN TRAJECTORY PROCESSING:
.....................................................
ACTION SETUP FOR PARM '6pti_ions.prmtop' (1 actions):
0: [pairdist out pairdist.dat mask :50<.2.4:WAT]
[:50<.2.4:WAT](12) [:50<.2.4:WAT](12)
----- product.nc (1-50000, 1) -----
0% 10% 20% 30% 40% 50% 60% 70% 80% 90% 100% Complete.
Read 50000 frames and processed 50000 frames.
TIME: Avg. throughput= 5261.8679 frames / second.
ACTION OUTPUT:
DATASETS (2 total):
PDIST_00001[Pr] "PDIST_00001[Pr]" (X-Y mesh), size is 493
PDIST_00001[std] "PDIST_00001[std]" (X-Y mesh), size is 493
DATAFILES (1 total):
pairdist.dat (Standard Data File): PDIST_00001[Pr] PDIST_00001[std]
RUN TIMING:
TIME: Init : 0.0000 s ( 0.00%)
TIME: Trajectory Process : 9.5023 s ( 99.99%)
TIME: Action Post : 0.0000 s ( 0.00%)
TIME: Analysis : 0.0000 s ( 0.00%)
TIME: Data File Write : 0.0006 s ( 0.01%)
TIME: Other : 0.0000 s ( 0.00%)
TIME: Run Total 9.5029 s
---------- RUN END ---------------------------------------------------
TIME: Total execution time: 9.5360 seconds.
--------------------------------------------------------------------------------
To cite CPPTRAJ use:
Daniel R. Roe and Thomas E. Cheatham, III, "PTRAJ and CPPTRAJ: Software for
Processing and Analysis of Molecular Dynamics Trajectory Data". J. Chem.
Theory Comput., 2013, 9 (7), pp 3084-3095.
Can anyone explain the aforementioned error message? And how to resolve the
issue?
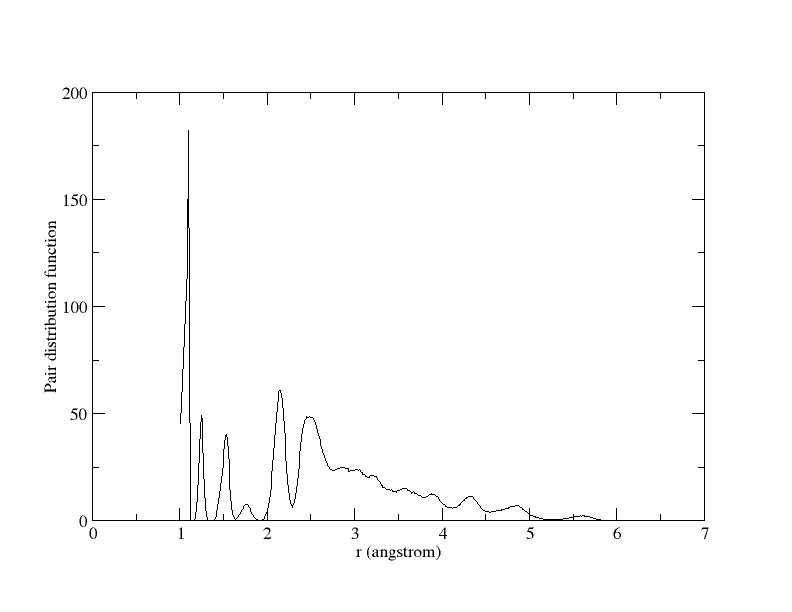
A plot of the pair distribution function is also attached.
Thanks in advance.
With regards,
Umesh
=======================================
Umesh C. Roy, Ph.D.
School of Computational and Integrative Sciences
Jawaharlal Nehru University, New Delhi
India-110067
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: pairdist.png)
