Date: Wed, 23 Dec 2020 19:49:34 +0530
Dear prof. Case,
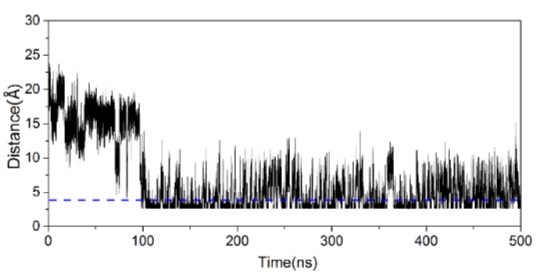
The cut off merely means that in the plot if the distance of 3.5 angstroms
or less is maintained, it was considered to be a good electrostatic
interaction in the system throughout the simulation. This notion was
considered referring some previous publications. Hope the attachment is now
visible to you.
On Wed, 23 Dec 2020 at 7:38 PM, David A Case <david.case.rutgers.edu> wrote:
> On Wed, Dec 23, 2020, Sruthi Sudhakar wrote:
>
> >I have been calculating the electrostatic distance plot between the
> >protonated nitrogen atom of my ligand and the negatively charged backbone
> >of DNA in a DNA-ligand complex. The distance was calculated using the
> >distance option in cpptraj. The protonated nitrogen belongs to the
> >pyrrodiline ring at the end of a propyl chain. The plot between the N+ and
> >OP is attached here. If the cut off of 3.5 angstroms is considered are
> the
> >high fluctuations (10-20 angstroms) seen in the plot possible or is it
> some
> >discrepancy? Kindly advise on the above.
>
> Nothing was attached...you might also be more specific about what you
> mean a "cut off of 3.5 angstroms"....
>
> ....dac
>
>
> _______________________________________________
> AMBER mailing list
> AMBER.ambermd.org
> http://lists.ambermd.org/mailman/listinfo/amber
>
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/jpeg attachment: IMG_3814.jpg)
