Date: Fri, 21 Feb 2020 10:48:32 +0900
Dear All (and David),
There are 7-CYS besides (CYS bound to HEM), none of these appear close to
any one of the CYS in pymol. Thus not sure if why this warning about CYS
being close should appear?
Which ambertools I should try on the rst7/pdb files, to find S-S distances
between these CYS to understand my parmed is giving the unexpected warning?
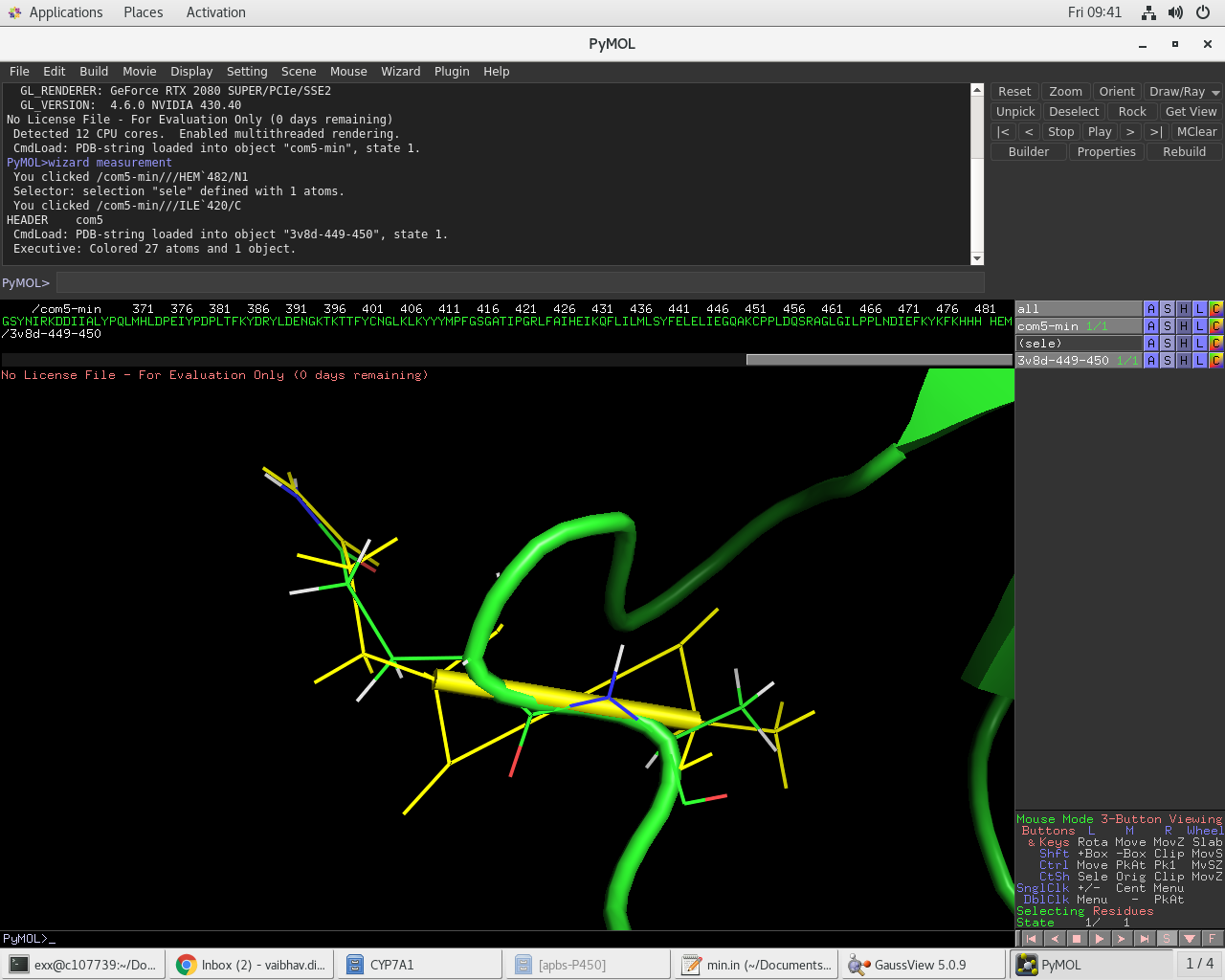
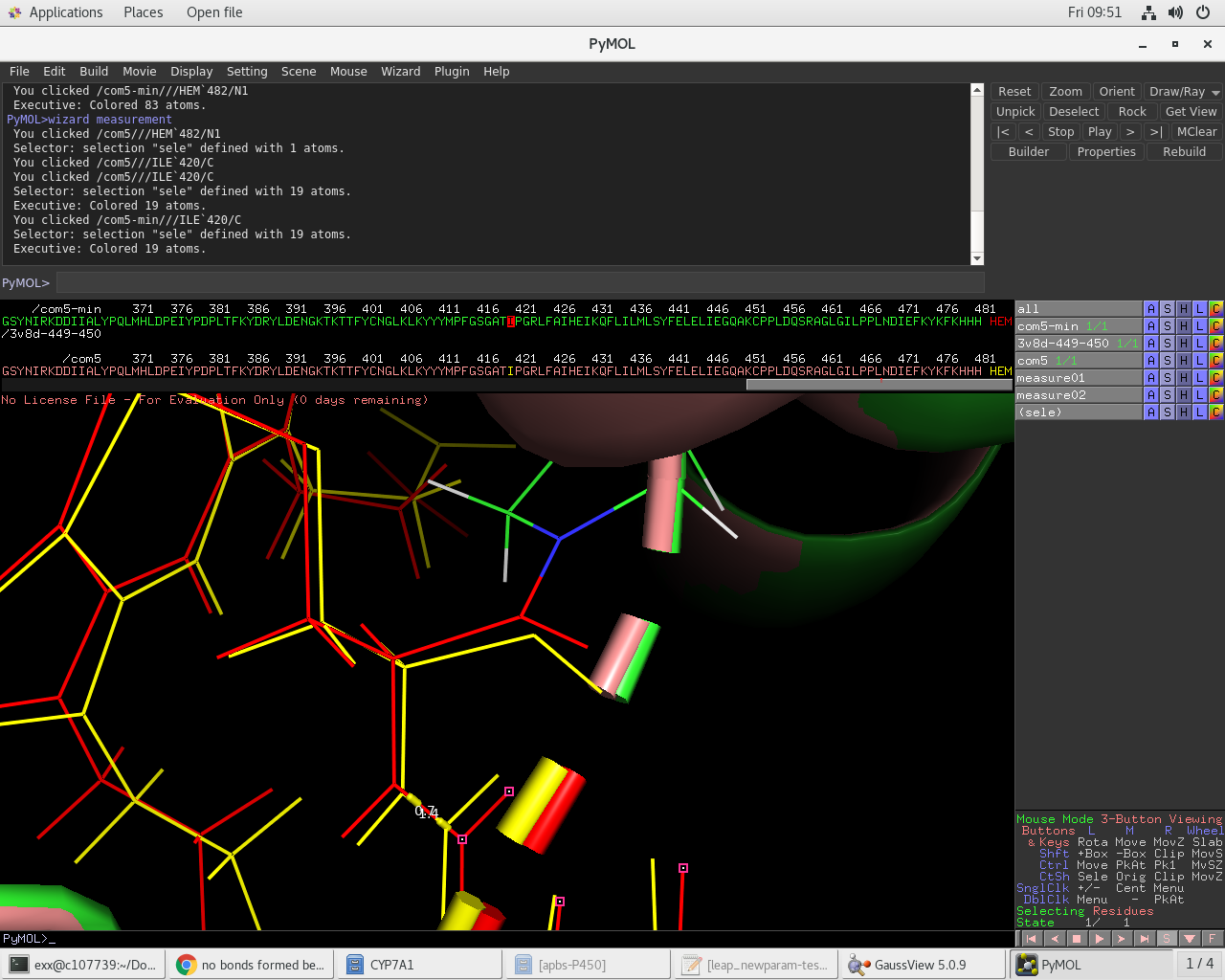
After minimization (element colors) the HEM to prot-backbone, and 449-450
amide long-bond has relaxed to expected distances as seen in the attached
images and parmed output.
thank you.
Loaded Amber topology file com5.prmtop with coordinates from
../md-CYP7A1/com5-min.rst7
Reading input from STDIN...
> printbonds :449 :450
Atom 1 Atom 2 R eq Frc Cnst Distance
Energy
7253 C ( C) 7255 N ( N) 1.3350 490.0000 1.3392
0.0085
> printbonds :420 :421
Atom 1 Atom 2 R eq Frc Cnst Distance
Energy
> printbonds :420 :482
Atom 1 Atom 2 R eq Frc Cnst Distance
Energy
6759 C ( C) 7783 N1 ( n) 1.3790 427.6000 1.3604
0.1482
> printbonds :421 :482
Atom 1 Atom 2 R eq Frc Cnst Distance
Energy
6761 N ( N) 7790 C3 ( c) 1.3790 427.6000 1.3916
0.0681
> quit
Done!
(base) [exx.c107739 xleap-mol2]$ grep CYS com5.pdb
ATOM 321 N CYS 21 13.410 9.130 -21.607 -0.4157 1.5500
N
ATOM 322 H CYS 21 13.852 8.424 -22.178 0.2719 1.3000
H
ATOM 323 CA CYS 21 12.400 8.699 -20.616 0.0213 1.7000
C
ATOM 324 HA CYS 21 11.544 9.373 -20.648 0.1124 1.3000
H
ATOM 325 CB CYS 21 11.922 7.297 -20.914 -0.1231 1.7000
C
ATOM 326 HB2 CYS 21 11.084 7.026 -20.272 0.1112 1.3000
H
ATOM 327 HB3 CYS 21 11.626 7.200 -21.958 0.1112 1.3000
H
ATOM 328 SG CYS 21 13.163 6.072 -20.641 -0.3119 1.8000
S
ATOM 329 HG CYS 21 12.411 5.032 -20.990 0.1933 0.8000
H
ATOM 330 C CYS 21 12.913 8.749 -19.175 0.5973 1.7000
C
ATOM 331 O CYS 21 14.112 8.691 -18.930 -0.5679 1.5000
O
ATOM 718 N CYS 46 21.744 -1.785 -26.749 -0.4157 1.5500
N
ATOM 719 H CYS 46 21.715 -1.445 -27.700 0.2719 1.3000
H
ATOM 720 CA CYS 46 20.630 -1.463 -25.866 0.0213 1.7000
C
ATOM 721 HA CYS 46 20.733 -2.007 -24.927 0.1124 1.3000
H
ATOM 722 CB CYS 46 20.605 0.045 -25.561 -0.1231 1.7000
C
ATOM 723 HB2 CYS 46 20.693 0.611 -26.488 0.1112 1.3000
H
ATOM 724 HB3 CYS 46 19.671 0.306 -25.064 0.1112 1.3000
H
ATOM 725 SG CYS 46 21.931 0.640 -24.482 -0.3119 1.8000
S
ATOM 726 HG CYS 46 21.550 1.914 -24.483 0.1933 0.8000
H
ATOM 727 C CYS 46 19.349 -1.828 -26.554 0.5973 1.7000
C
ATOM 728 O CYS 46 19.261 -1.758 -27.771 -0.5679 1.5000
O
ATOM 1076 N CYS 67 29.407 6.054 -44.104 -0.4157 1.5500
N
ATOM 1077 H CYS 67 29.897 5.500 -43.416 0.2719 1.3000
H
ATOM 1078 CA CYS 67 29.591 5.729 -45.509 0.0213 1.7000
C
ATOM 1079 HA CYS 67 29.665 6.642 -46.099 0.1124 1.3000
H
ATOM 1080 CB CYS 67 30.864 4.935 -45.687 -0.1231 1.7000
C
ATOM 1081 HB2 CYS 67 31.046 4.725 -46.741 0.1112 1.3000
H
ATOM 1082 HB3 CYS 67 31.717 5.472 -45.272 0.1112 1.3000
H
ATOM 1083 SG CYS 67 30.829 3.330 -44.855 -0.3119 1.8000
S
ATOM 1084 HG CYS 67 32.051 2.956 -45.223 0.1933 0.8000
H
ATOM 1085 C CYS 67 28.413 4.889 -46.019 0.5973 1.7000
C
ATOM 1086 O CYS 67 27.723 4.230 -45.235 -0.5679 1.5000
O
ATOM 2406 N CYS 152 11.381 33.634 -29.749 -0.4157 1.5500
N
ATOM 2407 H CYS 152 11.817 33.501 -28.848 0.2719 1.3000
H
ATOM 2408 CA CYS 152 11.962 32.932 -30.924 0.0213 1.7000
C
ATOM 2409 HA CYS 152 12.241 33.663 -31.682 0.1124 1.3000
H
ATOM 2410 CB CYS 152 13.218 32.128 -30.528 -0.1231 1.7000
C
ATOM 2411 HB2 CYS 152 13.013 31.511 -29.653 0.1112 1.3000
H
ATOM 2412 HB3 CYS 152 13.537 31.492 -31.354 0.1112 1.3000
H
ATOM 2413 SG CYS 152 14.639 33.140 -30.102 -0.3119 1.8000
S
ATOM 2414 HG CYS 152 15.458 32.126 -29.838 0.1933 0.8000
H
ATOM 2415 C CYS 152 10.953 32.019 -31.552 0.5973 1.7000
C
ATOM 2416 O CYS 152 10.859 31.919 -32.798 -0.5679 1.5000
O
ATOM 4989 N CYS 311 18.194 43.497 -48.588 -0.4157 1.5500
N
ATOM 4990 H CYS 311 18.722 44.355 -48.662 0.2719 1.3000
H
ATOM 4991 CA CYS 311 18.375 42.482 -49.620 0.0213 1.7000
C
ATOM 4992 HA CYS 311 17.807 41.590 -49.358 0.1124 1.3000
H
ATOM 4993 CB CYS 311 17.884 42.994 -50.991 -0.1231 1.7000
C
ATOM 4994 HB2 CYS 311 18.371 43.938 -51.237 0.1112 1.3000
H
ATOM 4995 HB3 CYS 311 18.102 42.260 -51.767 0.1112 1.3000
H
ATOM 4996 SG CYS 311 16.099 43.309 -51.083 -0.3119 1.8000
S
ATOM 4997 HG CYS 311 16.094 43.711 -52.351 0.1933 0.8000
H
ATOM 4998 C CYS 311 19.846 42.131 -49.707 0.5973 1.7000
C
ATOM 4999 O CYS 311 20.644 42.912 -50.230 -0.5679 1.5000
O
ATOM 6465 N CYS 402 41.103 13.935 -33.174 -0.4157 1.5500
N
ATOM 6466 H CYS 402 41.359 13.577 -34.083 0.2719 1.3000
H
ATOM 6467 CA CYS 402 40.868 12.983 -32.098 0.0213 1.7000
C
ATOM 6468 HA CYS 402 40.794 13.511 -31.148 0.1124 1.3000
H
ATOM 6469 CB CYS 402 39.569 12.219 -32.334 -0.1231 1.7000
C
ATOM 6470 HB2 CYS 402 38.722 12.902 -32.391 0.1112 1.3000
H
ATOM 6471 HB3 CYS 402 39.625 11.633 -33.251 0.1112 1.3000
H
ATOM 6472 SG CYS 402 39.185 11.062 -31.030 -0.3119 1.8000
S
ATOM 6473 HG CYS 402 38.051 10.619 -31.566 0.1933 0.8000
H
ATOM 6474 C CYS 402 42.039 12.007 -32.067 0.5973 1.7000
C
ATOM 6475 O CYS 402 42.305 11.329 -33.077 -0.5679 1.5000
O
ATOM 7287 N CYS 452 22.856 29.989 -17.585 -0.4157 1.5500
N
ATOM 7288 H CYS 452 23.792 29.989 -17.205 0.2719 1.3000
H
ATOM 7289 CA CYS 452 22.567 29.107 -18.729 0.0213 1.7000
C
ATOM 7290 HA CYS 452 22.005 29.660 -19.481 0.1124 1.3000
H
ATOM 7291 CB CYS 452 23.854 28.589 -19.365 -0.1231 1.7000
C
ATOM 7292 HB2 CYS 452 24.535 29.406 -19.604 0.1112 1.3000
H
ATOM 7293 HB3 CYS 452 24.358 27.875 -18.714 0.1112 1.3000
H
ATOM 7294 SG CYS 452 23.551 27.728 -20.906 -0.3119 1.8000
S
ATOM 7295 HG CYS 452 24.825 27.436 -21.155 0.1933 0.8000
H
ATOM 7296 C CYS 452 21.686 27.920 -18.316 0.5973 1.7000
C
ATOM 7297 O CYS 452 22.094 27.099 -17.477 -0.5679 1.5000
O
[image: image.png]
[image: image.png]
On Thu, Feb 20, 2020 at 10:50 PM David A Case <david.case.rutgers.edu>
wrote:
> On Thu, Feb 20, 2020, Vaibhav Dixit wrote:
>
> >I got the syntax right, but the rst7 file is still failing to load in vmd.
>
> Can't help with the vmd problem. You log file doesn't show any errors,
> and ends with the phrase
> "Info) Finished with coordinate file com5.rst7."
> So it's not clear what the failure is. But, again, I don't use vmd
> myself.
>
> >
> >MissingDisulfide: Detected two cysteine residues whose sulfur atoms are
> >within 3 Angstroms.j
>
> This warning is triggered because you have two CYS residues whose sulfur
> atoms are withing 3Ang, just like the message says. In your case, these
> are almost certainly the two cysteines bonded to Fe, and you know that
> they don't for a disulfide bond. So you can ignore this one.
>
> >
> >LongBondWarning: Atoms 7253 (GLN 449 [C]) and 7255 (ALA 450 [N]) are
> bonded
> > (equilibrium length 1.335 A) but are 4.826 A apart. This
> > often indicates gaps in the original sequence and should
> be
> > checked carefully.
>
> Sounds to me like a real problem that you should "check carefully", as
> the message indicates. Look at the header of your pdb file for
> information about missing residues, and (of course) visualize what is
> going on around residues 449 and 450.
>
> ...dac
>
>
-- Regards, Dr. Vaibhav A. Dixit, Visiting Scientist at the Manchester Institute of Biotechnology (MIB), The University of Manchester, 131 Princess Street, Manchester M1 7DN, UK. AND Assistant Professor, Department of Pharmacy, ▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄ Birla Institute of Technology and Sciences Pilani (BITS-Pilani), VidyaVihar Campus, street number 41, Pilani, Rajasthan 333031. India. Phone No. +91 1596 255652, Mob. No. +91-7709129400, Email: vaibhav.dixit.pilani.bits-pilani.ac.in, vaibhavadixit.gmail.com http://www.bits-pilani.ac.in/pilani/vaibhavdixit/profile https://www.linkedin.com/in/vaibhav-dixit-b1a07a39/ ORCID ID: https://orcid.org/0000-0003-4015-2941 http://scholar.google.co.in/citations?user=X876BKcAAAAJ&hl=en&oi=sra P Please consider the environment before printing this e-mail
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: image.png)
(image/png attachment: 02-image.png)
