Date: Sat, 15 Feb 2020 12:55:04 -0700
Dear everyone,
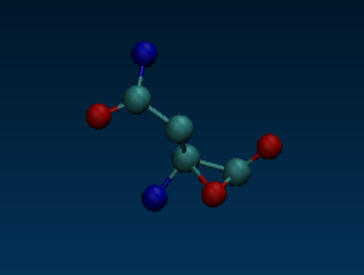
I got an error when using H++ to parameterize a protein-ligand binding
complex and I got the following error: "FATAL: Atom .R<NASN 107>.A<OXT 17>
does not have a type. ", which means that the OXT atom (the oxygen atom of
a terminal residue) of aspargine as the N-terminal residue (residue 107) of
the ligand does not have a type. Since the C-terminal residue (residue 106)
of the protein also has an OXT atom, but there was no issue with it, I
really don't know why I got an error like this. Another weird thing is that
an asparagine as an N-terminal residue should only have two oxygen atoms,
but there are three oxygen atoms in the residue. Using VMD to visualize the
structure, I also found that there is a weird/unreasonable bond between the
OXT atom and its adjacent atom, which seems to be wrong and should be
deleted. (The ASN residue was built by MODELLER since it was originally
missing, but I assume that the structure output by MODELLER should be
correct). I know that deleting the OXT atom would avoid the error that I
got from H++, but now I'm very confused about if I should delete it, since
from the visualization by VMD the OXT atom seemed wrong and should be
deleted, but deleting that atom also makes me feel unsafe as I thought I
should not even have gotten this error at the beginning. Does anyone have
suggestions for this? Thank you so much!
[image: image.png]
Best,
Wei-Tse
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: image.png)
