Date: Wed, 19 Jun 2019 14:11:48 +0100
Dear All,
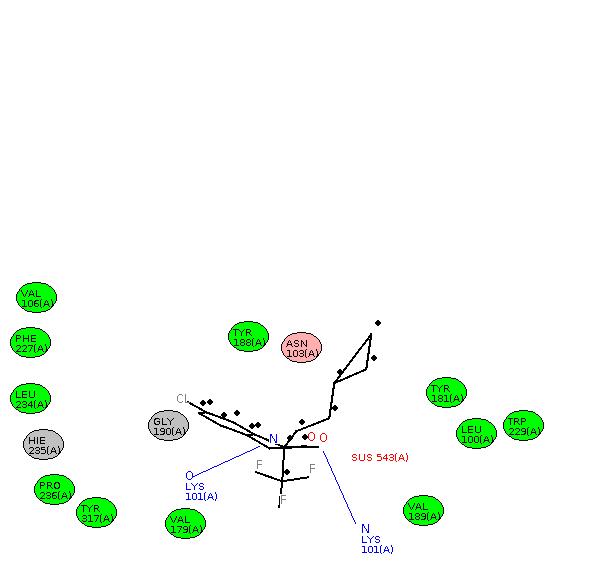
Am happy to share that I wrote a script to convert ambpdb pdb files into
RCSB-formatted files which LeView is recognizing and generating useful
(beautiful) 2D graphs of ligand-receptor interactions.
Example image generated from sustiva tutorial prmtop inpcrd files is
attached.
The script is taking 5min 8sec on a protein with 543 residues and thus I
think is not an optimum code, but does the job.
I believe one can use this to generate 2D picture from MD trajectories and
combine them into a cartoon animation (gif) of the MD simulation.
Please let me know if anyone is interested in utilizing this script along
with LeView.
FYI - LeView authors are cc'd (thanks to them for helping me fix the errors
that LeView reports with ambpdb generated pdb files).
Prof. Case, thanks for your valuable comments/suggestions on this and other
topics.
best regards
On Tue, Jun 4, 2019 at 2:12 PM Vaibhav Dixit <vaibhavadixit.gmail.com>
wrote:
> I ran ambpdb with -bres flag, but it doesn't generate chain id which the
> Leview expects (since it is present even in pdb with only one chain). I
> will try sed/awk to insert chain id and see if it fixes the problem.
> My intention here is to be able to generate gif animation by combining
> many images (derived from MD trajectories via pdb files) like the one I
> attached earlier.
> This will give a focused view on which residues come within interaction
> distance without having to load the whole trajectory or look/analyze into
> text files generated by cpptraj. I hope that makes sense to you (and others
> on the list).
> Below is the example where Leview works to rcsb file, but fails to
> recognize files generated by pdb (in this case probably because it lacks
> chain id information).
>
> C:\Users\r11831vd\Downloads\LeViewLine2.0\LeViewLine>java -jar
> LeViewLine.jar 2prg -r 1 1 A -flat
> downloading 2prg PDB entry
>
> C:\Users\r11831vd\Downloads\LeViewLine2.0\LeViewLine>java -jar
> LeViewLine.jar leview-test.pdb -r 473 473 -flat
> Exception in thread "main" java.lang.StringIndexOutOfBoundsException:
> String index out of range: 30
> at java.lang.String.charAt(Unknown Source)
> at File2.getMolecule(File2.java:445)
> at LeView.main(LeView.java:323)
>
> C:\Users\r11831vd\Downloads\LeViewLine2.0\LeViewLine>
>
>
> On Tue, Jun 4, 2019 at 1:11 PM David A Case <david.case.rutgers.edu>
> wrote:
>
>> On Tue, Jun 04, 2019, Vaibhav Dixit wrote:
>>
>> >I noticed that there are (small) differences in PDB file formats between
>> >RCSB and AMBER.
>>
>> You don't say what you mean by "AMBER". [Using ambpdb gets a PDB-format
>> file that follows closely the wwPDB standard. Use the "-bres" option to
>> get standard residue names. Residue numbers and chainID's tend to get
>> lost when running Amber simulations, but these can be retrieved via
>> add_pdb and the "-ext" option to ambpdb.]
>>
>> You also don't say what differences you find, or what specific
>> problems LeVIew has.
>>
>> >LeView 2D interaction map
>>
>> The hbond command in cpptraj reports on hydrogen bonds. The "closest"
>> command can provide contact information between the ligand and receptor.
>> But, by all means, Use LeView if it suits your needs.
>>
>> ....dac
>>
>>
>> _______________________________________________
>> AMBER mailing list
>> AMBER.ambermd.org
>> http://lists.ambermd.org/mailman/listinfo/amber
>>
>
>
> --
>
> Regards,
>
> Dr. Vaibhav A. Dixit,
>
> Visiting Scientist at the Manchester Institute of Biotechnology (MIB), The
> University of Manchester, 131 Princess Street, Manchester M1 7DN, UK.
> AND
> Assistant Professor,
> Department of Pharmacy,
> ▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄
> Birla Institute of Technology and Sciences Pilani (BITS-Pilani),
> VidyaVihar Campus, street number 41, Pilani, Rajasthan 333031.
> India.
> Phone No. +91 1596 255652, Mob. No. +91-7709129400,
> Email: vaibhav.dixit.pilani.bits-pilani.ac.in, vaibhavadixit.gmail.com
> http://www.bits-pilani.ac.in/pilani/vaibhavdixit/profile
> https://www.linkedin.com/in/vaibhav-dixit-b1a07a39/
>
> ORCID ID: https://orcid.org/0000-0003-4015-2941
>
> http://scholar.google.co.in/citations?user=X876BKcAAAAJ&hl=en&oi=sra
>
> P Please consider the environment before printing this e-mail
>
>
-- Regards, Dr. Vaibhav A. Dixit, Visiting Scientist at the Manchester Institute of Biotechnology (MIB), The University of Manchester, 131 Princess Street, Manchester M1 7DN, UK. AND Assistant Professor, Department of Pharmacy, ▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄ Birla Institute of Technology and Sciences Pilani (BITS-Pilani), VidyaVihar Campus, street number 41, Pilani, Rajasthan 333031. India. Phone No. +91 1596 255652, Mob. No. +91-7709129400, Email: vaibhav.dixit.pilani.bits-pilani.ac.in, vaibhavadixit.gmail.com http://www.bits-pilani.ac.in/pilani/vaibhavdixit/profile https://www.linkedin.com/in/vaibhav-dixit-b1a07a39/ ORCID ID: https://orcid.org/0000-0003-4015-2941 http://scholar.google.co.in/citations?user=X876BKcAAAAJ&hl=en&oi=sra P Please consider the environment before printing this e-mail
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
- text/plain attachment: 1FKO_trunc_sus_A0.txt
(image/jpeg attachment: sus-test-rcsb_A0.jpg)
