Date: Sun, 2 Jun 2019 10:07:29 +0100
Dear All,
I am heating up a minimized system (CYP450
<http://archive.ambermd.org/201905/0331.html>) in the implicit solvent with
473 residues.
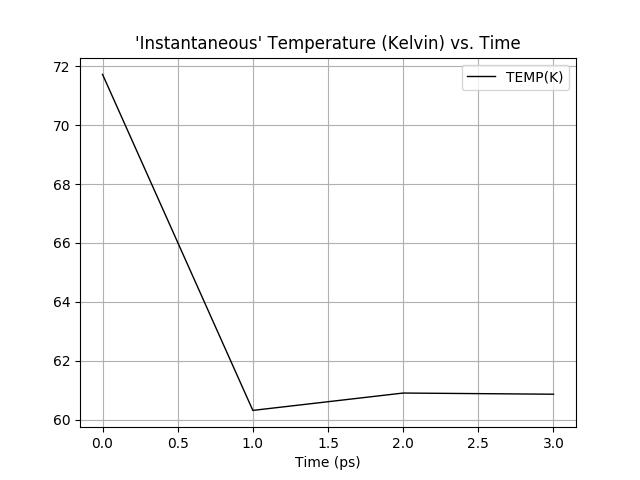
The mdout_analyzer.py heat.out graph attached shows that the temperature is
not rising above 60K. I took the input template from consph tutorial here
<http://ambermd.org/tutorials/advanced/tutorial18/section2.htm>.
I ran another longer simulation assuming that it would let the temp rise to
target 300 K, but this doesn't seem to have happened.
Can you please check/suggest what mistake am making, in either reading the
out file or setting the input files?
My PBS script is given below for reference.
I briefly read the "17.6.7. Temperature regulation" section, but not being
an expert couldn't quickly understand the reason for temp not moving above
60K.
[r11831vd.hlogin1 [csf3] 3A4-implicit-sol]$ tail heat2.out
NSTEP = 1500 TIME(PS) = 3.000 *TEMP(K) = 60.86* PRESS =
0.0
Etot = -13864.3602 EKtot = 1170.9430 EPtot =
-15035.3033
BOND = 516.2669 ANGLE = 1670.5681 DIHED =
4835.1562
1-4 NB = 1616.9575 1-4 EEL = 18935.0265 VDWAALS =
-4329.3383
EELEC = -31874.0368 EGB = -6477.3909 RESTRAINT =
71.4875
EAMBER (non-restraint) = -15106.7907
------------------------------------------------------------------------------
NMR restraints: Bond = 0.000 Angle = 0.000 Torsion = 0.000
===============================================================================
[r11831vd.hlogin1 [csf3] 3A4-implicit-sol]$
heat2.in
---------------------
Implicit solvent constant pH initial heating mdin
&cntrl
imin=0, irest=0, ntx=1,
ntpr=500, ntwx=500, nstlim=10000000,
dt=0.002, ntt=3, tempi=60,
temp0=300, tautp=2.0, ig=-1,
ntp=0, ntc=2, ntf=2, cut=30,
ntb=0, igb=2, tol=0.000001,
nrespa=1, saltcon=0.1, icnstph=1,
ntcnstph=100000000,
gamma_ln=5.0, ntwr=500, ioutfm=1,
nmropt=1, ntr=1, restraint_wt=2.0,
restraintmask='.CA,C,O,N',
/
&wt
TYPE='TEMP0', ISTEP1=1, ISTEP2=500000,
VALUE1=60.0, VALUE2=300.0,
/
&wt TYPE='END' /
PBS script
[r11831vd.hlogin1 [csf3] 3A4-implicit-sol]$ more amber-heat
#!/bin/bash --login
#$ -cwd
#$ -pe smp.pe 24
# Load the software
module load apps/intel-17.0/amber/18-at19-may2019
mpirun -n $NSLOTS sander.MPI -O -i heat2.in -o heat2.out -c
P450-1W0E-gas-heat.rst7 -r P450-1W0E-gas-heat2.rst7 -p P450-1W0E-gas.prmtop
-
ref P450-1W0E-gas-heat.rst7 -cpin P450-1W0E-gas-min.cpin -x
P450-1W0E-gas-heat2.nc -cpout P450-1W0E-gas-heat2.cpout
[r11831vd.hlogin1 [csf3] 3A4-implicit-sol]$
-- Regards, Dr. Vaibhav A. Dixit, Visiting Scientist at the Manchester Institute of Biotechnology (MIB), The University of Manchester, 131 Princess Street, Manchester M1 7DN, UK. AND Assistant Professor, Department of Pharmacy, ▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄▄ Birla Institute of Technology and Sciences Pilani (BITS-Pilani), VidyaVihar Campus, street number 41, Pilani, Rajasthan 333031. India. Phone No. +91 1596 255652, Mob. No. +91-7709129400, Email: vaibhav.dixit.pilani.bits-pilani.ac.in, vaibhavadixit.gmail.com http://www.bits-pilani.ac.in/pilani/vaibhavdixit/profile https://www.linkedin.com/in/vaibhav-dixit-b1a07a39/ ORCID ID: https://orcid.org/0000-0003-4015-2941 http://scholar.google.co.in/citations?user=X876BKcAAAAJ&hl=en&oi=sra P Please consider the environment before printing this e-mail
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: heat2.png)
