Date: Tue, 8 May 2018 12:04:21 +0530
Dear Amber users,
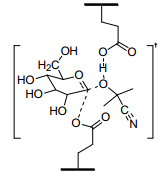
I am hoping to perform a SMD QM/MM for glucosidase-glucoside complex. I was
able to successfully locate and confirm TS for glucoside hydrolysis
reaction. My TS contains an active site residue intact. Hence i can't use
the TS as it is due to the presence of the same residue in original crystal
structure and i don't suppose it is a good approach to remove residue part
from TS because then it would change the TS entirely. I really appreciate
any suggestion or otherwise on this matter.
Furthermore I have attached my TS structure with this. How would you
recommend using RESP charge derived prmtop rather than using GLYCAM
forcefield (due to the deviation from standard carbohydrate structure) ?
Looking forward to hear your help.
Best Wishes,
D L S Dinuka
-- D L Senal Dinuka Grad.Chem., A.I.Chem.C. Research Assistant College of Chemical Sciences Institute of Chemistry Ceylon Rajagiriya Sri Lanka +94 77 627 4678
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: image.png)
