Date: Thu, 21 Dec 2017 17:36:13 +0500
Dear Amber Users,
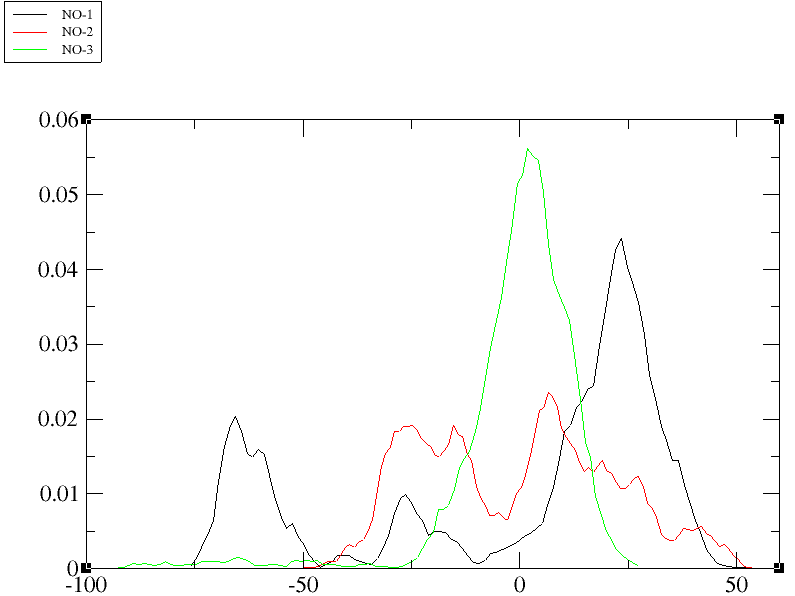
Here I am attaching the HIST .agr plot that represented three modes. These
modes not overlap properly. Is this plot have any meaning, I mean to
present in the paper or I need to run PCA analysis for calculating more
modes rather then three or do something other. Kindly guide thanks.
On Tue, Dec 19, 2017 at 7:24 PM, Rana Rehan Khalid <rrkhalid.umich.edu>
wrote:
> Yes sir It works now thank you.
>
> On Tue, Dec 19, 2017 at 7:23 PM, Daniel Roe <daniel.r.roe.gmail.com>
> wrote:
>
>> You should upgrade to AmberTools 17 on your cluster.
>>
>> -Dan
>>
>> On Mon, Dec 18, 2017 at 4:16 PM, Rana Rehan Khalid <rrkhalid.umich.edu>
>> wrote:
>> > Sir In my laptop i have Trajectory Analysis. V17.00, while on our
>> cluster
>> > we have amber 14 and cpptraj 14.
>> >
>> > On Mon, Dec 18, 2017 at 11:58 PM, Daniel Roe <daniel.r.roe.gmail.com>
>> wrote:
>> >
>> >> What version of cpptraj is on your laptop and what version is on the
>> >> cluster?
>> >>
>> >> -Dan
>> >>
>> >> On Sat, Dec 16, 2017 at 10:25 AM, Rana Rehan Khalid <
>> rrkhalid.umich.edu>
>> >> wrote:
>> >> > when I did the cpptraj pca analysis on my laptop and run the command
>> one
>> >> by
>> >> > one it works but when i submit my job with the above mentioned
>> script on
>> >> > cluster it show these warnings and job again stop. Thanks
>> >> >
>> >> > [diagmatrix oxxx-covar out oxxx-evecs.dat \ ]
>> >> > Warning: # of eigenvectors specified is < 1 (0) and 'thermo' not
>> >> specified.
>> >> > Warning: Specify # eigenvectors with 'vecs <#>'. Setting to 1.
>> >> > Changed DataFile 'oxxx-evecs.dat' type to Evecs file for set
>> >> > Modes_00004
>> >> > DIAGMATRIX: Diagonalizing matrix oxxx-covar and writing modes to
>> >> > oxxx-evecs.dat
>> >> > Calculating 1 eigenvectors
>> >> > TIME: Total execution time: 230.3207 seconds.
>> >> >
>> >> > On Sat, Dec 16, 2017 at 6:38 PM, Rana Rehan Khalid <
>> rrkhalid.umich.edu>
>> >> > wrote:
>> >> >
>> >> >> Hi sir Thank you. I understand now the point.I change the script as
>> you
>> >> >> guide and ran it works. kindly guide me if any thing wrong in it.
>> >> >>
>> >> >> Regards
>> >> >>
>> >> >> parm ox_solv.prmtop
>> >> >> trajin oxy_1.mdcrd
>> >> >> trajin oxy_2.mdcrd
>> >> >> trajin oxy_3.mdcrd
>> >> >> trajin oxy_4.mdcrd
>> >> >> trajin oxy_5.mdcrd
>> >> >> trajin oxy_6.mdcrd
>> >> >> trajin oxy_7.mdcrd
>> >> >> trajin oxy_8.mdcrd
>> >> >> trajin oxy_10.mdcrd
>> >> >> trajin oxy_11.mdcrd
>> >> >> rms first :1-189&!.H=
>> >> >> average crdset MyAverage
>> >> >> run
>> >> >> rms ref MyAverage :1-189&!.H=
>> >> >> matrix covar name bay1-covar :1-189&!.H=
>> >> >> diagmatrix bay1-covar out bbb-evecs.dat \
>> >> >> vecs 3 name myEvecs \
>> >> >> nmwiz nmwizvecs 3 nmwizfile bbb.nmd nmwizmask :1-189&!.H=
>> >> >> run
>> >> >> rms ref MyAverage :1-189&!.H=
>> >> >> projection HNOX-O2 evecs bbb-evecs.dat myEvecs beg 1 end 3
>> :1-189&!.H=
>> >> >> crdframes 1,70000
>> >> >> run
>> >> >> hist HNOX-O2:1 bins 100 out oxy-hist.agr norm name OXY-1
>> >> >> hist HNOX-O2:2 bins 100 out oxy-hist.agr norm name OXY-2
>> >> >> hist HNOX-O2:3 bins 100 out oxy-hist.agr norm name OXY-3
>> >> >> run
>> >> >> ##############
>> >> >> clear all
>> >> >> readdata bbb-evecs.dat name Evecs
>> >> >> parm ox_solv.prmtop
>> >> >> parmstrip !(:1-189&!.H=)
>> >> >> parmwrite out oxx-modes.prmtop
>> >> >> runanalysis modes name Evecs trajout oxx-mode1-10.nc \
>> >> >> pcmin -100 pcmax 100 tmode 1 trajoutmask :1-189&!.H= trajoutfmt
>> netcd
>> >> >>
>> >> >> On Sat, Dec 16, 2017 at 1:05 PM, Rana Rehan Khalid <
>> rrkhalid.umich.edu>
>> >> >> wrote:
>> >> >>
>> >> >>> Hi
>> >> >>> I am confused about the <option> I tried to compare it with other
>> PCA
>> >> >>> script and manual to get understanding but not get point. Is it
>> mode
>> >> data
>> >> >>> set like .dat Kindly guide me. Thanks
>> >> >>> diagmatrix bay1-covar name bay1-modes <options>
>> >> >>> modes name bay1-modes <options>
>> >> >>> projection MyProjection evecs bay1-modes <options>
>> >> >>>
>> >> >>> Regards
>> >> >>>
>> >> >>> On Thu, Dec 14, 2017 at 11:21 PM, Daniel Roe <
>> daniel.r.roe.gmail.com>
>> >> >>> wrote:
>> >> >>>
>> >> >>>> Hi,
>> >> >>>>
>> >> >>>> Try doing it without creating a COORDS data set - this stores all
>> your
>> >> >>>> coordinates in memory, which is convenient if you have the memory
>> to
>> >> >>>> spare, but in this case you do not. This means you need to modify
>> your
>> >> >>>> coordinates each time you process them (i.e. you need to rms-fit
>> >> >>>> before you project etc). Something like:
>> >> >>>>
>> >> >>>> # First run to generate average structure
>> >> >>>> trajin bay_1.mdcrd
>> >> >>>> ...
>> >> >>>> rms first <mask>
>> >> >>>> average crdset MyAverage
>> >> >>>> run
>> >> >>>> # Second run to generate covariance matrix and modes. Also run
>> modes
>> >> >>>> analysis.
>> >> >>>> rms ref MyAverage <mask>
>> >> >>>> matrix covar name bay1-covar <mask>
>> >> >>>> diagmatrix bay1-covar name bay1-modes <options>
>> >> >>>> modes name bay1-modes <options>
>> >> >>>> run
>> >> >>>> # Example third run for projection
>> >> >>>> rms ref MyAverage <mask>
>> >> >>>> projection MyProjection evecs bay1-modes <options>
>> >> >>>>
>> >> >>>> Hope this helps,
>> >> >>>>
>> >> >>>> -Dan
>> >> >>>>
>> >> >>>>
>> >> >>>>
>> >> >>>>
>> >> >>>> On Wed, Dec 13, 2017 at 4:58 PM, Rana Rehan Khalid <
>> >> rrkhalid.umich.edu>
>> >> >>>> wrote:
>> >> >>>> > Dear Amber users
>> >> >>>> >
>> >> >>>> > I am trying to run the PCA script every time job stop due to
>> this
>> >> >>>> exceeded
>> >> >>>> > limit of memory
>> >> >>>> >
>> >> >>>> > job 16802098 exceeded MEM usage hard limit (39282 > 2048)
>> >> >>>> >
>> >> >>>> > I use this script for running the job
>> >> >>>> > #!/bin/bash
>> >> >>>> > #PBS -l nodes=1:ppn=1
>> >> >>>> > #PBS -l walltime=100:00:00
>> >> >>>> > cd /nfs/amino-home/rrkhalid/bay60
>> >> >>>> > /nfs/amino-library/local/Amber14/bin/cpptraj -p bay.prmtop -i
>> >> >>>> pca.cpptraj
>> >> >>>> >
>> >> >>>> > and pca.cpptraj script is this each trajectory( 1-10 ) file run
>> >> 750000
>> >> >>>> > steps.
>> >> >>>> > rajin bay_1.mdcrd
>> >> >>>> > trajin bay_2.mdcrd
>> >> >>>> > trajin bay_3.mdcrd
>> >> >>>> > trajin bay_4.mdcrd
>> >> >>>> > trajin bay_5.mdcrd
>> >> >>>> > trajin bay_6.mdcrd
>> >> >>>> > trajin bay_7.mdcrd
>> >> >>>> > trajin bay_8.mdcrd
>> >> >>>> > trajin bay_9.mdcrd
>> >> >>>> > trajin bay_10.mdcrd
>> >> >>>> > rms first :1-189&!.H=
>> >> >>>> > average crdset average
>> >> >>>> > createcrd avgtrajectories
>> >> >>>> > run
>> >> >>>> > crdaction avgtrajectories rms ref average :1-189&!.H=
>> >> >>>> > crdaction avgtrajectories matrix covar name bay1-covar
>> :1-189&!.H=
>> >> >>>> > runanalysis diagmatrix bay1-covar out bay1-evecs.dat \
>> >> >>>> > vecs 3 name myEvecs \
>> >> >>>> > nmwiz nmwizvecs 3 nmwizfile bay.nmd nmwizmask :1-189&!.H=
>> >> >>>> > ##########
>> >> >>>> > clear all
>> >> >>>> > readdata bay1-evecs.dat name Evecs
>> >> >>>> > parm bay.prmtop
>> >> >>>> > parmstrip !(:1-189&!.H=)
>> >> >>>> > parmwrite out bay-modes.prmtop
>> >> >>>> > runanalysis modes name Evecs trajout bay-mode1.nc \
>> >> >>>> > pcmin -100 pcmax 100 tmode 1 trajoutmask :1-189&!.H=
>> trajoutfmt
>> >> netcd
>> >> >>>> >
>> >> >>>> > kindly guide Thanks
>> >> >>>> > _______________________________________________
>> >> >>>> > AMBER mailing list
>> >> >>>> > AMBER.ambermd.org
>> >> >>>> > http://lists.ambermd.org/mailman/listinfo/amber
>> >> >>>>
>> >> >>>>
>> >> >>>>
>> >> >>>> --
>> >> >>>> -------------------------
>> >> >>>> Daniel R. Roe
>> >> >>>> Laboratory of Computational Biology
>> >> >>>> National Institutes of Health, NHLBI
>> >> >>>> 5635 Fishers Ln, Rm T900
>> >> >>>> Rockville MD, 20852
>> >> >>>> https://www.lobos.nih.gov/lcb
>> >> >>>>
>> >> >>>> _______________________________________________
>> >> >>>> AMBER mailing list
>> >> >>>> AMBER.ambermd.org
>> >> >>>> http://lists.ambermd.org/mailman/listinfo/amber
>> >> >>>>
>> >> >>>
>> >> >>>
>> >> >>
>> >> > _______________________________________________
>> >> > AMBER mailing list
>> >> > AMBER.ambermd.org
>> >> > http://lists.ambermd.org/mailman/listinfo/amber
>> >>
>> >>
>> >>
>> >> --
>> >> -------------------------
>> >> Daniel R. Roe
>> >> Laboratory of Computational Biology
>> >> National Institutes of Health, NHLBI
>> >> 5635 Fishers Ln, Rm T900
>> >> Rockville MD, 20852
>> >> https://www.lobos.nih.gov/lcb
>> >>
>> >> _______________________________________________
>> >> AMBER mailing list
>> >> AMBER.ambermd.org
>> >> http://lists.ambermd.org/mailman/listinfo/amber
>> >>
>> > _______________________________________________
>> > AMBER mailing list
>> > AMBER.ambermd.org
>> > http://lists.ambermd.org/mailman/listinfo/amber
>>
>>
>>
>> --
>> -------------------------
>> Daniel R. Roe
>> Laboratory of Computational Biology
>> National Institutes of Health, NHLBI
>> 5635 Fishers Ln, Rm T900
>> Rockville MD, 20852
>> https://www.lobos.nih.gov/lcb
>>
>> _______________________________________________
>> AMBER mailing list
>> AMBER.ambermd.org
>> http://lists.ambermd.org/mailman/listinfo/amber
>>
>
>
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: pca.png)
