Date: Wed, 29 Nov 2017 18:50:12 +0800
Dear professor:
I want to calculate detalG of two reactions by building thermodynamic cycle as follows:
Cu2+(aq) + Ligand(chloroform)+ 2Cl-(aq) --> Cu(Ligand)Cl2(chloroform) detalG1
detalG3 deatalG4
Ni2+(aq) + Ligand(chloroform)+ 2Cl-(aq) --> Ni(Ligand)Cl2(chloroform) detalG2
As a result, detaldeatalG =detalG1 -detalG2 = detalG4-detalG3. I choose to calculate detalG3 or detalG4 by Gaussian Quadrature and soft-core potentials.
The .prmtop and .inpcrd file for calculating detalG4(or detalG3) are created as follows:
Firstly, setup the model of Cu(Ligand)Cl2(chloroform) (or Cu2+(aq)) and run 3ns MD simulations. Then,create the .prmtop and .inpcrd of Cu(Ligand)Cl2(chloroform) (or Cu2+(aq)) base on the final structure after 3ns MD. Similarly, create the .prmtop and .inpcrd of Ni(Ligand)Cl2(chloroform) (or Ni2+(aq)) by changing copper to nickel in the final structure.
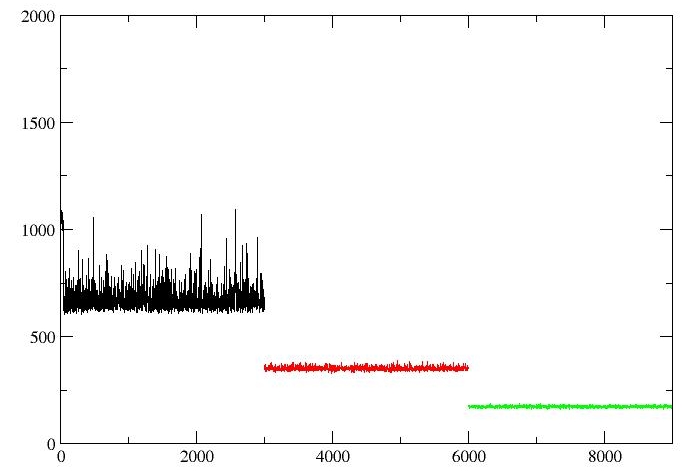
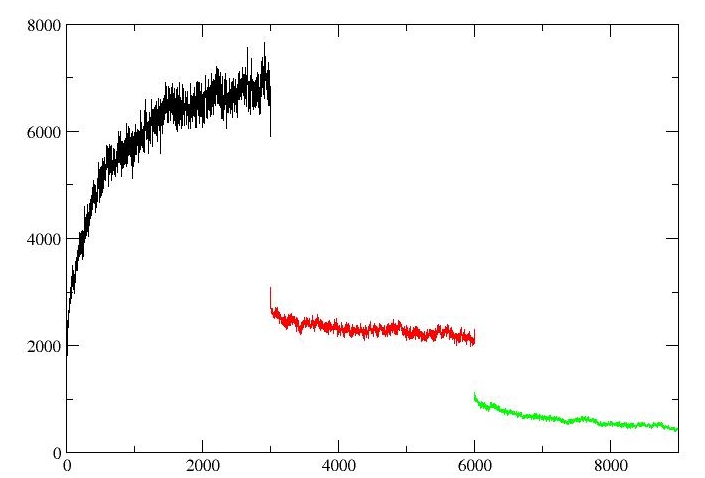
However, when I check for convergence for detalG4, the image is as follows(lambda=0.01592£¨black£©£¬lambda=0.08198£¨red£©£¬lambda=0.19331£¨green£©):
when I check for convergence for detalG3, the image is as follows(lambda=0.01592£¨black£©£¬lambda=0.08198£¨red£©£¬lambda=0.19331£¨green£©):
As shown in the figures, The value of dV/dL for the different lambda is not continuous. Is it correct? Whether there are problems in Figure2 or not.
If I do not describe the problem clearly, please tell me. I really wonder whether the steps I use to calculate detalG4(or detalG3) is right or not. Your any reply will help me a lot.
All the regards,
Yang Jinpeng
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/jpeg attachment: Catch.jpg)
(image/jpeg attachment: CatchAB46.jpg)
