Date: Thu, 19 Oct 2017 23:16:51 +0500
Hi Everybody
I saved one frame of my simulation in the pdb file. But when I open this
pdb file in the vmd, chimera and in pymol it does not show the bond between
Histidine and iron while in the .mdcrd file which i loaded after .prmtop
file shows all the heme pyrole bond and coordinate bond between histidine
and Fe. Similarly the bond between Fe and NO also can be seen in the
trajectory file but after saving the frame in to pdb it vanished. can you
please guide me how i can visualized the bond in vmd.
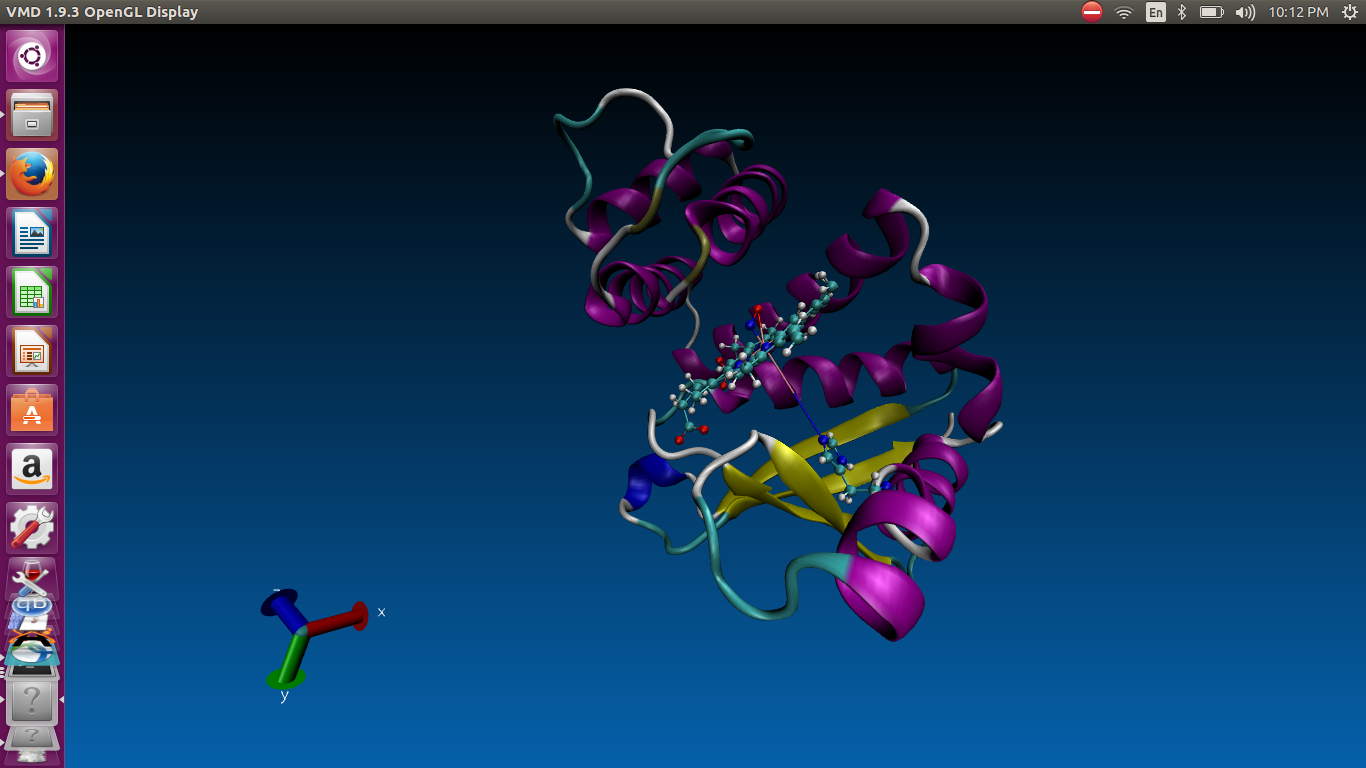
Here is the attached images 1st one is the captured during md run in this
image all bond shows
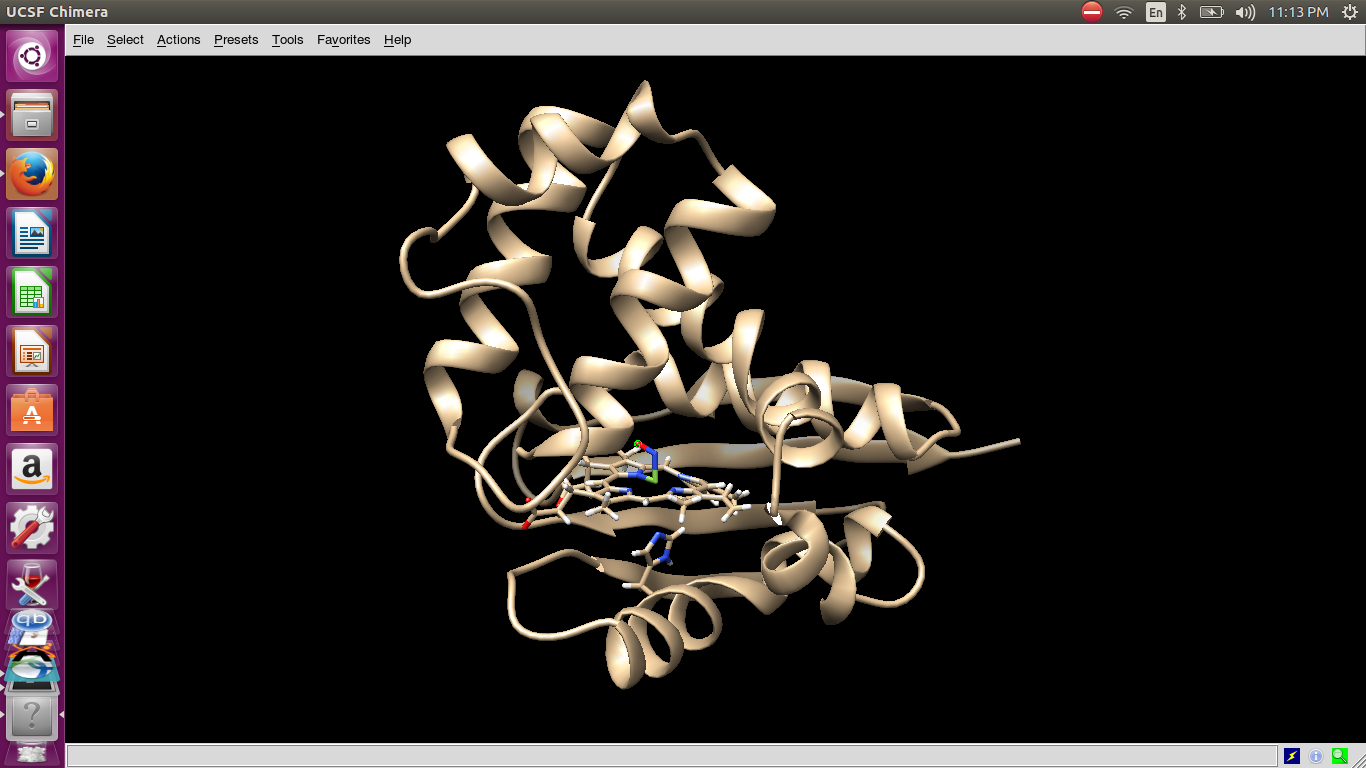
in the 2nd image is the pdb saved, one frame which i opened in chimera.
Regards
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: after_9_nano_sec.png)
(image/png attachment: Screenshot_from_2017-10-19_23-13-35.png)
