Date: Mon, 7 Aug 2017 21:49:45 -0400
Hello,
In the string methods, what does it means in the stsm.001.out file below:
# = NFE%STSM Initial
Pathway==================================================
# << anchor(1) : position = 3.960000, strength = 20.000000 >>
# << anchor(2) : position = -130.020000, strength = 20.000000 >>
#
-----------------------------------------------------------------------------
# MD time (ps), CV(1:2)
#
=============================================================================
0.0000 0.06914130 -2.26923847
0.0200 0.07271208 -2.73408209
0.0400 0.07652805 -2.84965414
0.0600 0.07795472 3.03822033
0.0800 0.08241867 3.12142878
0.1000 0.08495199 3.01791427
0.1200 0.08555569 2.86717038
.....
and
NFE : # << colvar(1) = 0.007826 0.022057 >>
NFE : # << colvar(2) = -2.694593 -2.964593 >>
NFE : # drifted center of image 1 : 40 0.00782640
-2.69459250
NFE : # drifted center of image 2 : 40 0.52288431
0.91052733
NFE : # drifted center of image 3 : 40 2.56373851
0.70046240
NFE : # drifted center of image 4 : 40 0.07509091
-2.20667518
NFE : # drifted center of image 5 : 40 0.89524373
-0.76143378
NFE : # smoothed center of image 1 : 40 0.00782640
-2.69459250
NFE : # smoothed center of image 2 : 40 0.57346627
0.89674353
NFE : # smoothed center of image 3 : 40 2.51138799
0.64872570
NFE : # smoothed center of image 4 : 40 0.06100224
-2.15863523
NFE : # smoothed center of image 5 : 40 0.88727440
-0.70817694
NFE : # smoothed center of image 6 : 40 0.72381698
-2.91261127
NFE : # reparametrized center of image 1 : 40 0.00782640
-2.69459250
NFE : # reparametrized center of image 2 : 40 0.42696645
1.12313090
NFE : # reparametrized center of image 3 : 40 1.98923377
0.73173851
NFE : # reparametrized center of image 4 : 40 1.92101428
0.24929126
NFE : # reparametrized center of image 5 : 40 0.12852031
-2.12319703
NFE : # reparametrized center of image 6 : 40 0.88562494
-0.71647809
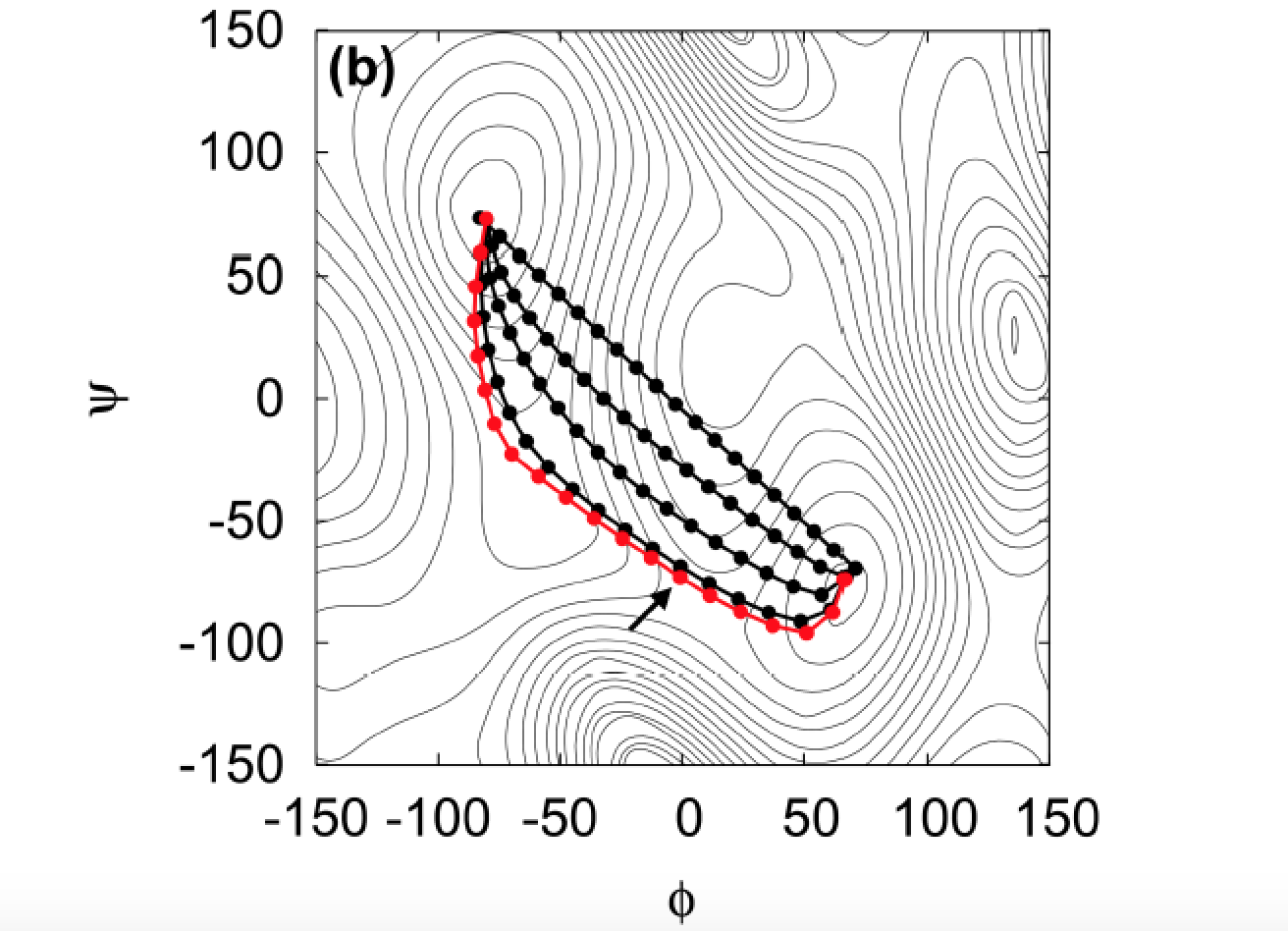
which one do I use to get the attached plot?
Below is my colvar file:
cv_file
&colvar ! Pseudo-dihedral
cv_type = 'COM_TORSION'
cv_ni = 57, cv_i =
138,139,141,143,144,147,148,149,200,201,203,204,205,206,209,210,212,213,422,423,425,426,430,431,432,434,435,486,487,489,490,491,492,493,495,496,499,500,0,156,157,158,0,188,189,190,0,168,169,171,172,176,177,178,180,181,0
anchor_position = 3.96
anchor_strength = 20.0
/
&colvar ! Glycosidic torsion
cv_type = 'TORSION'
cv_ni = 4, cv_i = 180, 168, 166, 165
anchor_position = -130.02
anchor_strength = 20.0
/
Best,
*Chunli*
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: Screenshot_2017-08-07_21.47.58.png)
