Date: Mon, 26 Sep 2016 19:06:02 +0800 (GMT+08:00)
Dear
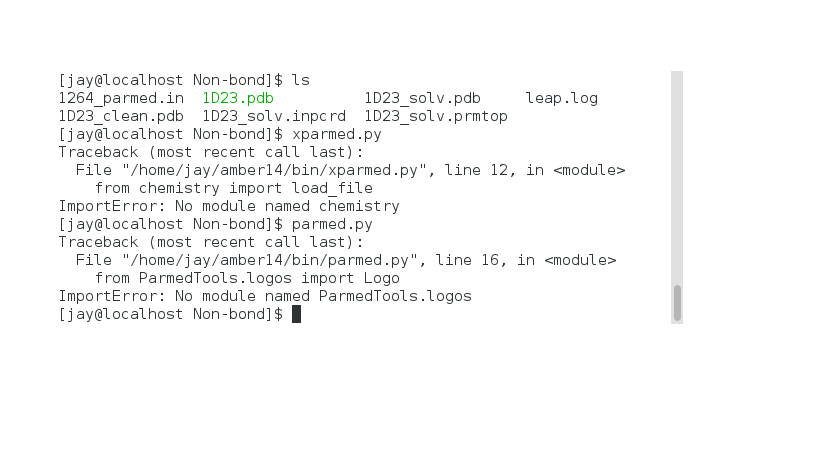
when I learned the tutorial20--Modeling a Magnesium-DNA System using the 12-6-4 LJ-Type Nonbonded Model. When inputting "Parmed" or "xParmed"according to the steps of the tutorial, I met this prolem as shown in the following picture:
The version of AMBER and AmberTool is 14 and 15, respectively. $AMBERHOME=/home/jay/amber14. How do I solve this problem?
Yang Jinpeng
sincerely
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: problem_when_using_ParmEd.png)
