Date: Fri, 8 Jul 2016 11:42:08 +0000 (UTC)
Dear zoran,
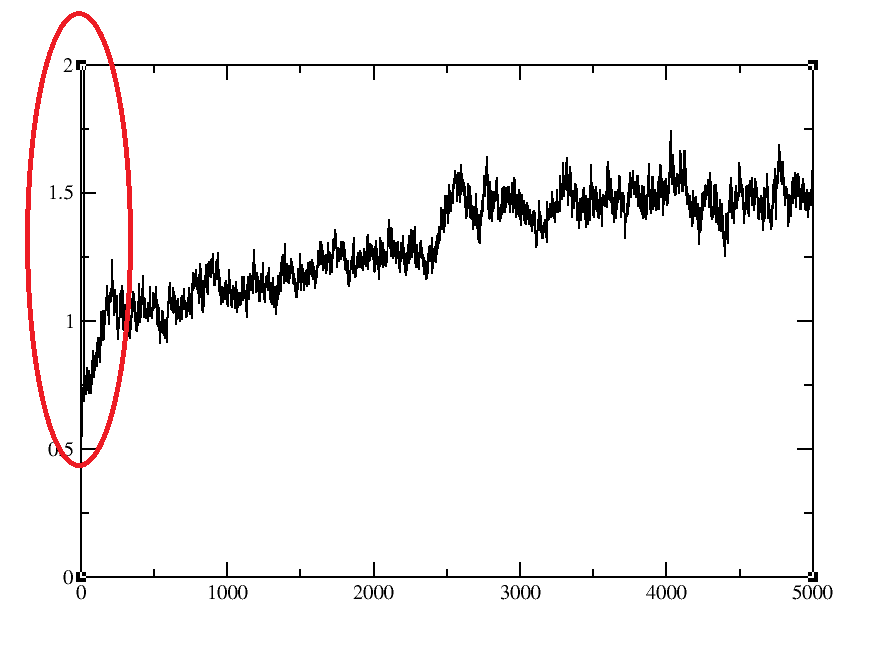
This rmsd plot is generated after 5ns production run. I have performed protein ligand complex md simulation. When I reimaged my mdcrd after production it gives me following error for 1ns trajectory; Warning: autoimage: Frame 20 imaging failed, box lengths are zero. While rest of proceed normally without this warning. you see in attached file that the rmsd jumps to near20 angstroms. This is a sign of something seriouslywrong with the trajectory.
Best Regards, Sarah
On Friday, July 8, 2016 4:17 AM, zoran matovic <zmatovic.kg.ac.rs> wrote:
Hi,
What kind of MD (heating, equil., production)? What kind of molecule is the
"complex"? What was the reference structure you have used for RMSD? What's
up to me you have nice equilibrium after 5 ns.
cheers
zoran
Dear all,
I have performed 5ns md simulation using amber14. Rmsd plot of one my
simulated complex shows unusua 20 angstrom fluctuation at starting ns. I am
unable to understand the reason. Rmsd plot is attached below. Kindly help
me. Best Regards, Sarah
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: rmsd.png)
