Date: Wed, 30 Mar 2016 12:49:34 +0500
Dear Daniel,
I have tried your readdata script again on my output files and one on my friend's output files. we both have same output files. readdata reads and lists all 24000 frames but plots only 12000 frames out of those 24000. following is the procedure I follow
>cpptraj
CPPTRAJ: Trajectory Analysis. V14.25
___ ___ ___ ___
| \/ | \/ | \/ |
_|_/\_|_/\_|_/\_|_
> readdata *.out name data
Reading '32a_prod1.out' as Amber MDOUT file with name 'data'
Reading from mdout files: 32a_prod1.out
Processing MDOUT: 32a_prod1.out
4000 frames
> readdata 32a_prod1.out name data
Reading '32a_prod1.out' as Amber MDOUT file with name 'data'
Reading from mdout files: 32a_prod1.out
Processing MDOUT: 32a_prod1.out
4000 frames
Warning: Appending to set 'data_Etot'. Using existing X step 0.5
data_Etot: 475.5 <= X <= 4475, 0.5
Warning: Appending to set 'data_EPtot'. Using existing X step 0.5
data_EPtot: 475.5 <= X <= 4475, 0.5
Warning: Appending to set 'data_BOND'. Using existing X step 0.5
data_BOND: 475.5 <= X <= 4475, 0.5
Warning: Appending to set 'data_ANGLE'. Using existing X step 0.5
data_ANGLE: 475.5 <= X <= 4475, 0.5
Warning: Appending to set 'data_DIHED'. Using existing X step 0.5
data_DIHED: 475.5 <= X <= 4475, 0.5
Warning: Appending to set 'data_VDW'. Using existing X step 0.5
data_VDW: 475.5 <= X <= 4475, 0.5
Warning: Appending to set 'data_EELEC'. Using existing X step 0.5
data_EELEC: 475.5 <= X <= 4475, 0.5
Warning: Appending to set 'data_VDW1-4'. Using existing X step 0.5
data_VDW1-4: 475.5 <= X <= 4475, 0.5
Warning: Appending to set 'data_EEL1-4'. Using existing X step 0.5
data_EEL1-4: 475.5 <= X <= 4475, 0.5
Warning: Appending to set 'data_RST'. Using existing X step 0.5
data_RST: 475.5 <= X <= 4475, 0.5
Warning: Appending to set 'data_Density'. Using existing X step 0.5
data_Density: 475.5 <= X <= 4475, 0.5
Warning: Appending to set 'data_EKtot'. Using existing X step 0.5
data_EKtot: 475.5 <= X <= 4475, 0.5
> readdata 32a_prod2.out name data
Reading '32a_prod2.out' as Amber MDOUT file with name 'data'
Reading from mdout files: 32a_prod2.out
Processing MDOUT: 32a_prod2.out
4000 frames
Warning: Appending to set 'data_Etot'. Using existing X step 0.5
data_Etot: 475.5 <= X <= 6475, 0.5
Warning: Appending to set 'data_EPtot'. Using existing X step 0.5
data_EPtot: 475.5 <= X <= 6475, 0.5
Warning: Appending to set 'data_BOND'. Using existing X step 0.5
data_BOND: 475.5 <= X <= 6475, 0.5
Warning: Appending to set 'data_ANGLE'. Using existing X step 0.5
data_ANGLE: 475.5 <= X <= 6475, 0.5
Warning: Appending to set 'data_DIHED'. Using existing X step 0.5
data_DIHED: 475.5 <= X <= 6475, 0.5
Warning: Appending to set 'data_VDW'. Using existing X step 0.5
data_VDW: 475.5 <= X <= 6475, 0.5
Warning: Appending to set 'data_EELEC'. Using existing X step 0.5
data_EELEC: 475.5 <= X <= 6475, 0.5
Warning: Appending to set 'data_VDW1-4'. Using existing X step 0.5
data_VDW1-4: 475.5 <= X <= 6475, 0.5
Warning: Appending to set 'data_EEL1-4'. Using existing X step 0.5
data_EEL1-4: 475.5 <= X <= 6475, 0.5
Warning: Appending to set 'data_RST'. Using existing X step 0.5
data_RST: 475.5 <= X <= 6475, 0.5
Warning: Appending to set 'data_Density'. Using existing X step 0.5
data_Density: 475.5 <= X <= 6475, 0.5
Warning: Appending to set 'data_EKtot'. Using existing X step 0.5
data_EKtot: 475.5 <= X <= 6475, 0.5
> readdata 32a_prod3.out name data
Reading '32a_prod3.out' as Amber MDOUT file with name 'data'
Reading from mdout files: 32a_prod3.out
Processing MDOUT: 32a_prod3.out
4000 frames
Warning: Appending to set 'data_Etot'. Using existing X step 0.5
data_Etot: 475.5 <= X <= 8475, 0.5
Warning: Appending to set 'data_EPtot'. Using existing X step 0.5
data_EPtot: 475.5 <= X <= 8475, 0.5
Warning: Appending to set 'data_BOND'. Using existing X step 0.5
data_BOND: 475.5 <= X <= 8475, 0.5
Warning: Appending to set 'data_ANGLE'. Using existing X step 0.5
data_ANGLE: 475.5 <= X <= 8475, 0.5
Warning: Appending to set 'data_DIHED'. Using existing X step 0.5
data_DIHED: 475.5 <= X <= 8475, 0.5
Warning: Appending to set 'data_VDW'. Using existing X step 0.5
data_VDW: 475.5 <= X <= 8475, 0.5
Warning: Appending to set 'data_EELEC'. Using existing X step 0.5
data_EELEC: 475.5 <= X <= 8475, 0.5
Warning: Appending to set 'data_VDW1-4'. Using existing X step 0.5
data_VDW1-4: 475.5 <= X <= 8475, 0.5
Warning: Appending to set 'data_EEL1-4'. Using existing X step 0.5
data_EEL1-4: 475.5 <= X <= 8475, 0.5
Warning: Appending to set 'data_RST'. Using existing X step 0.5
data_RST: 475.5 <= X <= 8475, 0.5
Warning: Appending to set 'data_Density'. Using existing X step 0.5
data_Density: 475.5 <= X <= 8475, 0.5
Warning: Appending to set 'data_EKtot'. Using existing X step 0.5
data_EKtot: 475.5 <= X <= 8475, 0.5
> readdata 32a_prod4.out name data
Reading '32a_prod4.out' as Amber MDOUT file with name 'data'
Reading from mdout files: 32a_prod4.out
Processing MDOUT: 32a_prod4.out
4000 frames
Warning: Appending to set 'data_Etot'. Using existing X step 0.5
data_Etot: 475.5 <= X <= 10475, 0.5
Warning: Appending to set 'data_EPtot'. Using existing X step 0.5
data_EPtot: 475.5 <= X <= 10475, 0.5
Warning: Appending to set 'data_BOND'. Using existing X step 0.5
data_BOND: 475.5 <= X <= 10475, 0.5
Warning: Appending to set 'data_ANGLE'. Using existing X step 0.5
data_ANGLE: 475.5 <= X <= 10475, 0.5
Warning: Appending to set 'data_DIHED'. Using existing X step 0.5
data_DIHED: 475.5 <= X <= 10475, 0.5
Warning: Appending to set 'data_VDW'. Using existing X step 0.5
data_VDW: 475.5 <= X <= 10475, 0.5
Warning: Appending to set 'data_EELEC'. Using existing X step 0.5
data_EELEC: 475.5 <= X <= 10475, 0.5
Warning: Appending to set 'data_VDW1-4'. Using existing X step 0.5
data_VDW1-4: 475.5 <= X <= 10475, 0.5
Warning: Appending to set 'data_EEL1-4'. Using existing X step 0.5
data_EEL1-4: 475.5 <= X <= 10475, 0.5
Warning: Appending to set 'data_RST'. Using existing X step 0.5
data_RST: 475.5 <= X <= 10475, 0.5
Warning: Appending to set 'data_Density'. Using existing X step 0.5
data_Density: 475.5 <= X <= 10475, 0.5
Warning: Appending to set 'data_EKtot'. Using existing X step 0.5
data_EKtot: 475.5 <= X <= 10475, 0.5
> readdata 32a_prod5.out name data
Reading '32a_prod5.out' as Amber MDOUT file with name 'data'
Reading from mdout files: 32a_prod5.out
Processing MDOUT: 32a_prod5.out
4000 frames
Warning: Appending to set 'data_Etot'. Using existing X step 0.5
data_Etot: 475.5 <= X <= 12475, 0.5
Warning: Appending to set 'data_EPtot'. Using existing X step 0.5
data_EPtot: 475.5 <= X <= 12475, 0.5
Warning: Appending to set 'data_BOND'. Using existing X step 0.5
data_BOND: 475.5 <= X <= 12475, 0.5
Warning: Appending to set 'data_ANGLE'. Using existing X step 0.5
data_ANGLE: 475.5 <= X <= 12475, 0.5
Warning: Appending to set 'data_DIHED'. Using existing X step 0.5
data_DIHED: 475.5 <= X <= 12475, 0.5
Warning: Appending to set 'data_VDW'. Using existing X step 0.5
data_VDW: 475.5 <= X <= 12475, 0.5
Warning: Appending to set 'data_EELEC'. Using existing X step 0.5
data_EELEC: 475.5 <= X <= 12475, 0.5
Warning: Appending to set 'data_VDW1-4'. Using existing X step 0.5
data_VDW1-4: 475.5 <= X <= 12475, 0.5
Warning: Appending to set 'data_EEL1-4'. Using existing X step 0.5
data_EEL1-4: 475.5 <= X <= 12475, 0.5
Warning: Appending to set 'data_RST'. Using existing X step 0.5
data_RST: 475.5 <= X <= 12475, 0.5
Warning: Appending to set 'data_Density'. Using existing X step 0.5
data_Density: 475.5 <= X <= 12475, 0.5
Warning: Appending to set 'data_EKtot'. Using existing X step 0.5
data_EKtot: 475.5 <= X <= 12475, 0.5
> list dataset
DATASETS:
12 data sets:
data[Etot] "data_Etot" (double), size is 24000
data[EPtot] "data_EPtot" (double), size is 24000
data[BOND] "data_BOND" (double), size is 24000
data[ANGLE] "data_ANGLE" (double), size is 24000
data[DIHED] "data_DIHED" (double), size is 24000
data[VDW] "data_VDW" (double), size is 24000
data[EELEC] "data_EELEC" (double), size is 24000
data[VDW1-4] "data_VDW1-4" (double), size is 24000
data[EEL1-4] "data_EEL1-4" (double), size is 24000
data[RST] "data_RST" (double), size is 24000
data[Density] "data_Density" (double), size is 24000
data[EKtot] "data_EKtot" (double), size is 24000
> writedata energy_tot.agr data[Etot]
data_Etot
> writedata energy_full.agr data[*]
data_Etot data_EPtot data_BOND data_ANGLE data_DIHED data_VDW data_EELEC data_VDW1-4 data_EEL1-4 data_RST data_Density data_EKtot
> quit
TIME: Total execution time: 132.9724 seconds.
--------------------------------------------------------------------------------
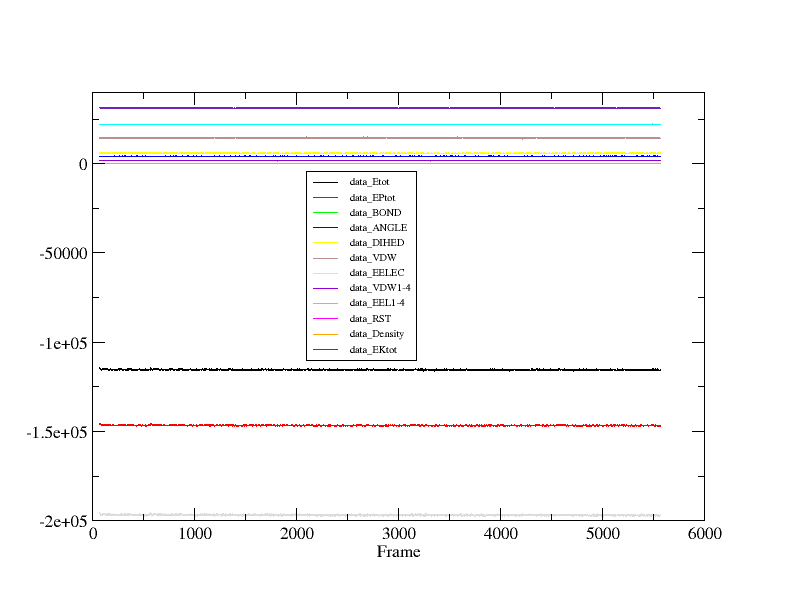
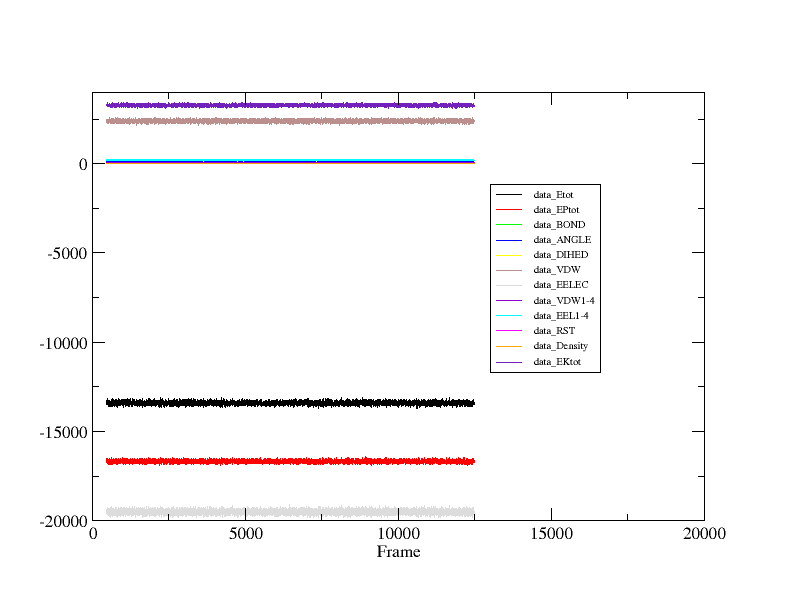
I have also attach the picture of my incomplete plot and complete plot of my friend's output files. Please guide whats wrong with this while it reads all 24000 frames of my output files. Where is the mistake which I am unable to understand....
Regards,
Sehrish Naz
Jr. Research Fellow,
Computational Chemistry Unit.
Dr. Panjwani Center for Molecular Medicine and Drug Research,
International Center for Chemical and Biological Sciences,
University of Karachi, Karachi-75270.
E-mail: Sehrish.naz.outlook.com
From: Sehrish Naz Aijaz<mailto:sehrish.naz.outlook.com>
Sent: 29 March 2016 09:03
To: Daniel Roe<mailto:daniel.r.roe.gmail.com>; AMBER Mailing List<mailto:amber.ambermd.org>
Subject: Re: [AMBER] problem in plotting total energy graphs by perl script
Sorry to say but I did not receive any mail from your side that’s why I again ask for help...
Sent from Mail<https://go.microsoft.com/fwlink/?LinkId=550986> for Windows 10
From: Daniel Roe<mailto:daniel.r.roe.gmail.com>
Sent: 28 March 2016 18:37
To: Sehrish Naz Aijaz<mailto:sehrish.naz.outlook.com>; AMBER Mailing List<mailto:amber.ambermd.org>
Subject: Re: [AMBER] problem in plotting total energy graphs by perl script
I responded to you 3 days ago (http://archive.ambermd.org/201603/0505.html).
Did you not receive my reply?
On Monday, March 28, 2016, Sehrish Naz Aijaz <sehrish.naz.outlook.com>
wrote:
> Dear all,
>
> Please suggest me I am waiting for some reply I am not getting that error
> regarding readdata what to do next its not taking my output files I
> guess....
>
>
>
> Sent from Mail <https://go.microsoft.com/fwlink/?LinkId=550986> for
> Windows 10
>
>
>
> *From: *Sehrish Naz Aijaz
> <javascript:_e(%7B%7D,'cvml','sehrish.naz.outlook.com');>
> *Sent: *25 March 2016 16:18
> *To: *Bill Ross <javascript:_e(%7B%7D,'cvml','ross.cgl.ucsf.edu');>; AMBER
> Mailing List <javascript:_e(%7B%7D,'cvml','amber.ambermd.org');>
> *Subject: *Re: [AMBER] problem in plotting total energy graphs by perl
> script
>
>
> Dear Bill,
> I have tried as you said to try just one output file but again its giving
> the following error as,
> writedata enegy.agr 32a_prod5.out
> Writing sets to enegy.agr, format 'Grace File'
> Warning: '32a_prod5.out' selects no data sets.
> Warning: 32a_prod5.out does not correspond to any data sets.
>
> Warning: File 'enegy.agr' has no sets containing data.
>
> And same error message was shown for the command
> Writedata energy.agr 32a_prod5.out_*
>
> Now what to do next please guide me....
>
> Regards,
> Sehrish
>
> Sent from Mail<https://go.microsoft.com/fwlink/?LinkId=550986> for
> Windows 10
>
> From: Bill Ross<mailto:ross.cgl.ucsf.edu
> <javascript:_e(%7B%7D,'cvml','ross.cgl.ucsf.edu');>>
> Sent: 25 March 2016 10:41
> To: AMBER Mailing List<mailto:amber.ambermd.org
> <javascript:_e(%7B%7D,'cvml','amber.ambermd.org');>>
> Subject: Re: [AMBER] problem in plotting total energy graphs by perl script
>
> And
>
> > writedata energy.agr 32a_prod5.out_*
>
> On 3/24/16 10:39 PM, Bill Ross wrote:
> > Just for fun, please try
> >
> > > writedata
> >
> > and
> >
> > > writedata energy.agr 32a_prod5.out
> >
> > Bill
> >
> > On 3/24/16 9:59 PM, Sehrish Naz Aijaz wrote:
> >> Dear Daniel,
> >> I try to run readdata commands as you mentioned. In the beginning it
> was going well as follows,
> >> CPPTRAJ: Trajectory Analysis. V15.00
> >> ___ ___ ___ ___
> >> | \/ | \/ | \/ |
> >> _|_/\_|_/\_|_/\_|_
> >>
> >> | Date/time: 03/25/16 09:52:32
> >> | Available memory: 550.168 MB
> >>
> >> Loading previous history from log 'cpptraj.log'
> >>> readdata *.out
> >> Reading '32a_prod1.out' as Amber MDOUT file with name
> '32a_prod1.out'
> >> Reading from mdout file: 32a_prod1.out
> >> 4000 frames
> >> Reading '32a_prod2.out' as Amber MDOUT file with name
> '32a_prod2.out'
> >> Reading from mdout file: 32a_prod2.out
> >> 4000 frames
> >> Reading '32a_prod3.out' as Amber MDOUT file with name
> '32a_prod3.out'
> >> Reading from mdout file: 32a_prod3.out
> >> 4000 frames
> >> Reading '32a_prod4.out' as Amber MDOUT file with name
> '32a_prod4.out'
> >> Reading from mdout file: 32a_prod4.out
> >> 4000 frames
> >> Reading '32a_prod5.out' as Amber MDOUT file with name
> '32a_prod5.out'
> >> Reading from mdout file: 32a_prod5.out
> >> 4000 frames
> >>> list dataset
> >> DATASETS:
> >> 75 data sets:
> >> 32a_prod1.out[Etot] "32a_prod1.out_Etot" (double), size is
> 4000
> >> 32a_prod1.out[EPtot] "32a_prod1.out_EPtot" (double), size is
> 4000
> >> 32a_prod1.out[BOND] "32a_prod1.out_BOND" (double), size is
> 4000
> >> 32a_prod1.out[ANGLE] "32a_prod1.out_ANGLE" (double), size is
> 4000
> >> 32a_prod1.out[DIHED] "32a_prod1.out_DIHED" (double), size is
> 4000
> >> 32a_prod1.out[VDW] "32a_prod1.out_VDW" (double), size is 4000
> >> 32a_prod1.out[EELEC] "32a_prod1.out_EELEC" (double), size is
> 4000
> >> 32a_prod1.out[VDW1-4] "32a_prod1.out_VDW1-4" (double), size
> is 4000
> >> 32a_prod1.out[EEL1-4] "32a_prod1.out_EEL1-4" (double), size
> is 4000
> >> 32a_prod1.out[RST] "32a_prod1.out_RST" (double), size is 4000
> >> 32a_prod1.out[Density] "32a_prod1.out_Density" (double), size
> is 4000
> >> 32a_prod1.out[EKtot] "32a_prod1.out_EKtot" (double), size is
> 4000
> >> 32a_prod1.out[VOLUME] "32a_prod1.out_VOLUME" (double), size
> is 4000
> >> 32a_prod1.out[TEMP] "32a_prod1.out_TEMP" (double), size is
> 4000
> >> 32a_prod1.out[PRESS] "32a_prod1.out_PRESS" (double), size is
> 4000
> >> 32a_prod2.out[Etot] "32a_prod2.out_Etot" (double), size is
> 4000
> >> 32a_prod2.out[EPtot] "32a_prod2.out_EPtot" (double), size is
> 4000
> >> 32a_prod2.out[BOND] "32a_prod2.out_BOND" (double), size is
> 4000
> >> 32a_prod2.out[ANGLE] "32a_prod2.out_ANGLE" (double), size is
> 4000
> >> 32a_prod2.out[DIHED] "32a_prod2.out_DIHED" (double), size is
> 4000
> >> 32a_prod2.out[VDW] "32a_prod2.out_VDW" (double), size is 4000
> >> 32a_prod2.out[EELEC] "32a_prod2.out_EELEC" (double), size is
> 4000
> >> 32a_prod2.out[VDW1-4] "32a_prod2.out_VDW1-4" (double), size
> is 4000
> >> 32a_prod2.out[EEL1-4] "32a_prod2.out_EEL1-4" (double), size
> is 4000
> >> 32a_prod2.out[RST] "32a_prod2.out_RST" (double), size is 4000
> >> 32a_prod2.out[Density] "32a_prod2.out_Density" (double), size
> is 4000
> >> 32a_prod2.out[EKtot] "32a_prod2.out_EKtot" (double), size is
> 4000
> >> 32a_prod2.out[VOLUME] "32a_prod2.out_VOLUME" (double), size
> is 4000
> >> 32a_prod2.out[TEMP] "32a_prod2.out_TEMP" (double), size is
> 4000
> >> 32a_prod2.out[PRESS] "32a_prod2.out_PRESS" (double), size is
> 4000
> >> 32a_prod3.out[Etot] "32a_prod3.out_Etot" (double), size is
> 4000
> >> 32a_prod3.out[EPtot] "32a_prod3.out_EPtot" (double), size is
> 4000
> >> 32a_prod3.out[BOND] "32a_prod3.out_BOND" (double), size is
> 4000
> >> 32a_prod3.out[ANGLE] "32a_prod3.out_ANGLE" (double), size is
> 4000
> >> 32a_prod3.out[DIHED] "32a_prod3.out_DIHED" (double), size is
> 4000
> >> 32a_prod3.out[VDW] "32a_prod3.out_VDW" (double), size is 4000
> >> 32a_prod3.out[EELEC] "32a_prod3.out_EELEC" (double), size is
> 4000
> >> 32a_prod3.out[VDW1-4] "32a_prod3.out_VDW1-4" (double), size
> is 4000
> >> 32a_prod3.out[EEL1-4] "32a_prod3.out_EEL1-4" (double), size
> is 4000
> >> 32a_prod3.out[RST] "32a_prod3.out_RST" (double), size is 4000
> >> 32a_prod3.out[Density] "32a_prod3.out_Density" (double), size
> is 4000
> >> 32a_prod3.out[EKtot] "32a_prod3.out_EKtot" (double), size is
> 4000
> >> 32a_prod3.out[VOLUME] "32a_prod3.out_VOLUME" (double), size
> is 4000
> >> 32a_prod3.out[TEMP] "32a_prod3.out_TEMP" (double), size is
> 4000
> >> 32a_prod3.out[PRESS] "32a_prod3.out_PRESS" (double), size is
> 4000
> >> 32a_prod4.out[Etot] "32a_prod4.out_Etot" (double), size is
> 4000
> >> 32a_prod4.out[EPtot] "32a_prod4.out_EPtot" (double), size is
> 4000
> >> 32a_prod4.out[BOND] "32a_prod4.out_BOND" (double), size is
> 4000
> >> 32a_prod4.out[ANGLE] "32a_prod4.out_ANGLE" (double), size is
> 4000
> >> 32a_prod4.out[DIHED] "32a_prod4.out_DIHED" (double), size is
> 4000
> >> 32a_prod4.out[VDW] "32a_prod4.out_VDW" (double), size is 4000
> >> 32a_prod4.out[EELEC] "32a_prod4.out_EELEC" (double), size is
> 4000
> >> 32a_prod4.out[VDW1-4] "32a_prod4.out_VDW1-4" (double), size
> is 4000
> >> 32a_prod4.out[EEL1-4] "32a_prod4.out_EEL1-4" (double), size
> is 4000
> >> 32a_prod4.out[RST] "32a_prod4.out_RST" (double), size is 4000
> >> 32a_prod4.out[Density] "32a_prod4.out_Density" (double), size
> is 4000
> >> 32a_prod4.out[EKtot] "32a_prod4.out_EKtot" (double), size is
> 4000
> >> 32a_prod4.out[VOLUME] "32a_prod4.out_VOLUME" (double), size
> is 4000
> >> 32a_prod4.out[TEMP] "32a_prod4.out_TEMP" (double), size is
> 4000
> >> 32a_prod4.out[PRESS] "32a_prod4.out_PRESS" (double), size is
> 4000
> >> 32a_prod5.out[Etot] "32a_prod5.out_Etot" (double), size is
> 4000
> >> 32a_prod5.out[EPtot] "32a_prod5.out_EPtot" (double), size is
> 4000
> >> 32a_prod5.out[BOND] "32a_prod5.out_BOND" (double), size is
> 4000
> >> 32a_prod5.out[ANGLE] "32a_prod5.out_ANGLE" (double), size is
> 4000
> >> 32a_prod5.out[DIHED] "32a_prod5.out_DIHED" (double), size is
> 4000
> >> 32a_prod5.out[VDW] "32a_prod5.out_VDW" (double), size is 4000
> >> 32a_prod5.out[EELEC] "32a_prod5.out_EELEC" (double), size is
> 4000
> >> 32a_prod5.out[VDW1-4] "32a_prod5.out_VDW1-4" (double), size
> is 4000
> >> 32a_prod5.out[EEL1-4] "32a_prod5.out_EEL1-4" (double), size
> is 4000
> >> 32a_prod5.out[RST] "32a_prod5.out_RST" (double), size is 4000
> >> 32a_prod5.out[Density] "32a_prod5.out_Density" (double), size
> is 4000
> >> 32a_prod5.out[EKtot] "32a_prod5.out_EKtot" (double), size is
> 4000
> >> 32a_prod5.out[VOLUME] "32a_prod5.out_VOLUME" (double), size
> is 4000
> >> 32a_prod5.out[TEMP] "32a_prod5.out_TEMP" (double), size is
> 4000
> >> 32a_prod5.out[PRESS] "32a_prod5.out_PRESS" (double), size is
> 4000
> >> but when I type the command;
> >> writedata energy.agr *.out
> >> it gave the following error;
> >> writedata energy.agr *.out
> >> Writing sets to energy.agr, format 'Grace File'
> >> Warning: '*.out' selects no data sets.
> >> Warning: *.out does not correspond to any data sets.
> >>
> >> Warning: File 'energy.agr' has no sets containing data.
> >> where I am doing mistake???
> >> I try to give all output files in writedata commamd as,
> >>> writedata energy.agr 32a_prod1.out 32a_prod2.out 32a_prod3.out
> 32a_prod4.out 32a_prod5.out
> >> Writing sets to energy.agr, format 'Grace File'
> >> Warning: '32a_prod1.out' selects no data sets.
> >> Warning: 32a_prod1.out does not correspond to any data sets.
> >> Warning: '32a_prod2.out' selects no data sets.
> >> Warning: 32a_prod2.out does not correspond to any data sets.
> >> Warning: '32a_prod3.out' selects no data sets.
> >> Warning: 32a_prod3.out does not correspond to any data sets.
> >> Warning: '32a_prod4.out' selects no data sets.
> >> Warning: 32a_prod4.out does not correspond to any data sets.
> >> Warning: '32a_prod5.out' selects no data sets.
> >> Warning: 32a_prod5.out does not correspond to any data sets.
> >>
> >> Warning: File 'energy.agr' has no sets containing data.
> >> again getting the same error. Please suggest me where am I doing
> mistake??
> >>
> >>
> >> Regards,
> >> Sehrish Naz
> >>
> >>
> >>
> >> Sent from Mail<https://go.microsoft.com/fwlink/?LinkId=550986> for
> Windows 10
> >>
> >> From: Daniel Roe<mailto:daniel.r.roe.gmail.com
> <javascript:_e(%7B%7D,'cvml','daniel.r.roe.gmail.com');>>
> >> Sent: 22 March 2016 21:01
> >> Cc: AMBER Mailing List<mailto:amber.ambermd.org
> <javascript:_e(%7B%7D,'cvml','amber.ambermd.org');>>
> >> Subject: Re: [AMBER] problem in plotting total energy graphs by perl
> script
> >>
> >> On Tue, Mar 22, 2016 at 9:50 AM, Sehrish Naz Aijaz
> >> <sehrish.naz.outlook.com
> <javascript:_e(%7B%7D,'cvml','sehrish.naz.outlook.com');>> wrote:
> >>> Can you please tell me the command you are suggesting because I have
> heard
> >>> this command first time please guide me.
> >> Assuming you have cpptraj installed you can do it interactively. Here
> >> is an example.
> >>
> >> $ cpptraj
> >> > readdata md.out
> >> Reading 'md.out' as Amber MDOUT file with name 'md.out'
> >> Reading from mdout file: md.out
> >> 100000 frames
> >> > list dataset
> >>
> >> DATASETS (14 total):
> >> md.out[Etot] "md.out_Etot" (double), size is 100000
> >> md.out[EPtot] "md.out_EPtot" (double), size is 100000
> >> md.out[BOND] "md.out_BOND" (double), size is 100000
> >> md.out[ANGLE] "md.out_ANGLE" (double), size is 100000
> >> md.out[DIHED] "md.out_DIHED" (double), size is 100000
> >> md.out[VDW] "md.out_VDW" (double), size is 100000
> >> md.out[EELEC] "md.out_EELEC" (double), size is 100000
> >> md.out[VDW1-4] "md.out_VDW1-4" (double), size is 100000
> >> md.out[EEL1-4] "md.out_EEL1-4" (double), size is 100000
> >> md.out[RST] "md.out_RST" (double), size is 100000
> >> md.out[EAMBER] "md.out_EAMBER" (double), size is 100000
> >> md.out[EKtot] "md.out_EKtot" (double), size is 100000
> >> md.out[TEMP] "md.out_TEMP" (double), size is 100000
> >> md.out[PRESS] "md.out_PRESS" (double), size is 100000
> >> > writedata energy.agr md.out[*]
> >> Writing sets to energy.agr, format 'Grace File'
> >> md.out_Etot md.out_EPtot md.out_BOND md.out_ANGLE md.out_DIHED
> >> md.out_VDW md.out_EELEC md.out_VDW1-4 md.out_EEL1-4 md.out_RST
> >> md.out_EAMBER md.out_EKtot md.out_TEMP md.out_PRESS
> >> > quit
> >>
> >> Then the file energy.agr will contain all of your energy data sets. If
> >> you just wanted e.g. the total potential energy you would use
> >> 'writedata eptot.agr md.out[EPtot]' instead.
> >>
> >> Hope this helps.
> >>
> >> -Dan
> >>
> >>> Thanks,
> >>>
> >>>
> >>>
> >>> Sent from Mail for Windows 10
> >>>
> >>>
> >>>
> >>> From: Daniel Roe
> >>> Sent: 21 March 2016 19:38
> >>> To: AMBER Mailing List
> >>>
> >>>
> >>> Subject: Re: [AMBER] problem in plotting total energy graphs by perl
> script
> >>>
> >>>
> >>>
> >>> Hi,
> >>>
> >>> FYI you can also use cpptraj to extract energies from MDOUT files via
> >>> the 'readdata' command. You can then analyze the data or write it out
> >>> into any format supported by cpptraj.
> >>>
> >>> -Dan
> >>>
> >>>
> >>> On Sun, Mar 20, 2016 at 10:56 PM, Sehrish Naz Aijaz
> >>> <sehrish.naz.outlook.com
> <javascript:_e(%7B%7D,'cvml','sehrish.naz.outlook.com');>> wrote:
> >>>> Dear Bill,
> >>>> Are you asking about mden content in my output files??? Or you are
> saying
> >>>> to use mden file generated during production runs???
> >>>> Can you tell me how can I plot those mden file which scripts will
> help for
> >>>> plotting and processing mden files???
> >>>>
> >>>> Sent from Mail<https://go.microsoft.com/fwlink/?LinkId=550986> for
> Windows
> >>>> 10
> >>>>
> >>>> From: Bill Ross<mailto:ross.cgl.ucsf.edu
> <javascript:_e(%7B%7D,'cvml','ross.cgl.ucsf.edu');>>
> >>>> Sent: 20 March 2016 01:11
> >>>> To: AMBER Mailing List<mailto:amber.ambermd.org
> <javascript:_e(%7B%7D,'cvml','amber.ambermd.org');>>
> >>>> Subject: Re: [AMBER] problem in plotting total energy graphs by perl
> >>>> script
> >>>>
> >>>> Have you looked at mden file contents? They could be easier to work
> with.
> >>>>
> >>>> Bill
> >>>>
> >>>> On 3/19/16 3:16 AM, Sehrish Naz Aijaz wrote:
> >>>>> Dear Hai,
> >>>>> I try to run this in my amber12 but it gives the following error.
> Can you
> >>>>> explain this to me...
> >>>>> Traceback (most recent call last):
> >>>>> File "/usr/local/amber14/bin/mdout_analyzer.py", line 3, in
> <module>
> >>>>> from tkFileDialog import askopenfilenames
> >>>>> ImportError: No module named tkFileDialog
> >>>>>
> >>>>> Sent from Mail<https://go.microsoft.com/fwlink/?LinkId=550986> for
> >>>>> Windows 10
> >>>>>
> >>>>> From: Hai Nguyen<mailto:nhai.qn.gmail.com
> <javascript:_e(%7B%7D,'cvml','nhai.qn.gmail.com');>>
> >>>>> Sent: 19 March 2016 14:48
> >>>>> To: AMBER Mailing List<mailto:amber.ambermd.org
> <javascript:_e(%7B%7D,'cvml','amber.ambermd.org');>>
> >>>>> Subject: Re: [AMBER] problem in plotting total energy graphs by perl
> >>>>> script
> >>>>>
> >>>>> I think it should be compatible (Jason might correct me if I am
> wrong).
> >>>>>
> >>>>> Here is the tutorial:
> http://jswails.wikidot.com/helpful-scripts#toc8
> >>>>>
> >>>>> Cheers
> >>>>> Hai
> >>>>>
> >>>>> On Sat, Mar 19, 2016 at 2:26 AM, Sehrish Naz Aijaz
> >>>>> <sehrish.naz.outlook.com
> <javascript:_e(%7B%7D,'cvml','sehrish.naz.outlook.com');>>
> >>>>> wrote:
> >>>>>
> >>>>>> Dear Hai,
> >>>>>> I have amber12 in my system. Is this script compatible for amber12
> which
> >>>>>> I
> >>>>>> think not compatible with amber12. So what should I do???
> >>>>>>
> >>>>>> Sent from Mail<https://go.microsoft.com/fwlink/?LinkId=550986> for
> >>>>>> Windows 10
> >>>>>>
> >>>>>> From: Hai Nguyen<mailto:nhai.qn.gmail.com
> <javascript:_e(%7B%7D,'cvml','nhai.qn.gmail.com');>>
> >>>>>> Sent: 19 March 2016 13:56
> >>>>>> To: AMBER Mailing List<mailto:amber.ambermd.org
> <javascript:_e(%7B%7D,'cvml','amber.ambermd.org');>>
> >>>>>> Subject: Re: [AMBER] problem in plotting total energy graphs by perl
> >>>>>> script
> >>>>>>
> >>>>>> Can you try to use mdout_analyzer.py in AmberTools15? (It's pain to
> >>>>>> debug
> >>>>>> Perl code)
> >>>>>>
> >>>>>> Check http://ambermd.org/doc12/Amber15.pdf (page 515)
> >>>>>>
> >>>>>> Hai
> >>>>>>
> >>>>>> On Sat, Mar 19, 2016 at 1:25 AM, Sehrish Naz Aijaz <
> >>>>>> sehrish.naz.outlook.com
> <javascript:_e(%7B%7D,'cvml','sehrish.naz.outlook.com');>>
> >>>>>> wrote:
> >>>>>>
> >>>>>>> Dear all,
> >>>>>>> I had run 10 ns simulation (AMBER12) of large compounds in TIP3P
> box
> >>>>>>> to
> >>>>>>> check their stable conformation with time. the input file I used
> for
> >>>>>>> production is
> >>>>>>> NPT production with no restrains
> >>>>>>> &cntrl
> >>>>>>> imin=0, ntx=7, irest=1, ntrx=1, ntxo=1,
> >>>>>>> ntpr=500, ntwx=500, ntwv=200, ntwe=200,
> >>>>>>> ntf=2, ntb=2, cut=10.0,
> >>>>>>> nsnb=100, igb=0,
> >>>>>>> nstlim=2000000,
> >>>>>>> t=0.0, dt=0.001,
> >>>>>>> ntt=3, gamma_ln=1.0, tempi=300.0, temp0=300.0,
> >>>>>>> vlimit=20,
> >>>>>>> ntp=1, taup=1.0, pres0=1.0, comp=44.6,
> >>>>>>> ntc=2, tol=0.00000001,
> >>>>>>> /
> >>>>>>>
> >>>>>>> after getting the production files. I tried to calculate the total
> >>>>>>> energy
> >>>>>>> graphs of the system by using perl script as follows
> >>>>>>> . #!/usr/bin/perl
> >>>>>>>
> >>>>>>>
> >>>>>>> if ($#ARGV < 0) {
> >>>>>>> print " Incorrect usage...\n";
> >>>>>>> exit;
> >>>>>>> }
> >>>>>>>
> >>>>>>>
> >>>>>>> foreach $i ( 0..$#ARGV ) {
> >>>>>>> $filein = $ARGV[$i];
> >>>>>>> $checkfile = $filein;
> >>>>>>> $checkfile =~ s/\.Z//;
> >>>>>>> if ( $filein ne $checkfile ) {
> >>>>>>> open(INPUT, "zcat $filein |") ||
> >>>>>>> die "Cannot open compressed $filein -- $!\n";
> >>>>>>> } else {
> >>>>>>> open(INPUT, $filein) || die "Cannot open $filein -- $!\n";
> >>>>>>> }
> >>>>>>> print "Processing sander output file ($filein)...\n";
> >>>>>>> &process_input;
> >>>>>>> close(INPUT);
> >>>>>>> }
> >>>>>>>
> >>>>>>> print "Starting output...\n";
> >>>>>>> .sortedkeys = sort by_number keys(%TIME);
> >>>>>>> .sortedavgkeys = sort by_number keys(%AVG_TIME);
> >>>>>>>
> >>>>>>> foreach $i ( TEMP, TSOLUTE, TSOLVENT, PRES, EKCMT, ETOT, EKTOT,
> EPTOT,
> >>>>>>> DENSITY, VOLUME, ESCF ) {
> >>>>>>> print "Outputing summary.$i\n";
> >>>>>>> open(OUTPUT, "> summary.$i");
> >>>>>>> %outarray = eval "\%$i";
> >>>>>>> foreach $j ( .sortedkeys ) {
> >>>>>>> print OUTPUT "$j ", $outarray{$j}, "\n";
> >>>>>>> }
> >>>>>>> close (OUTPUT);
> >>>>>>>
> >>>>>>> print "Outputing summary_avg.$i\n";
> >>>>>>> open(OUTPUT, "> summary_avg.$i");
> >>>>>>> %outarray = eval "\%AVG_$i";
> >>>>>>> foreach $j ( .sortedavgkeys ) {
> >>>>>>> print OUTPUT "$j ", $outarray{$j}, "\n";
> >>>>>>> }
> >>>>>>> close (OUTPUT);
> >>>>>>>
> >>>>>>> print "Outputing summary_rms.$i\n";
> >>>>>>> open(OUTPUT, "> summary_rms.$i");
> >>>>>>> %outarray = eval "\%RMS_$i";
> >>>>>>> foreach $j ( .sortedavgkeys ) {
> >>>>>>> print OUTPUT "$j ", $outarray{$j}, "\n";
> >>>>>>> }
> >>>>>>> close (OUTPUT);
> >>>>>>>
> >>>>>>>
> >>>>>>> }
> >>>>>>>
> >>>>>>>
> >>>>>>> sub by_number {
> >>>>>>> if ($a < $b) {
> >>>>>>> -1;
> >>>>>>> } elsif ($a == $b) {
> >>>>>>> 0;
> >>>>>>> } elsif ($a > $b) {
> >>>>>>> 1;
> >>>>>>> }
> >>>>>>> }
> >>>>>>>
> >>>>>>> sub process_input {
> >>>>>>>
> >>>>>>> $status = 0;
> >>>>>>> $debug = 0;
> >>>>>>> while ( <INPUT> ) {
> >>>>>>> $string = $_;
> >>>>>>>
> >>>>>>> print $_ if ( ! /NB-upda/ && $debug );
> >>>>>>>
> >>>>>>> if (/A V E R A G E S/) {
> >>>>>>> $averages = 1;
> >>>>>>> ($averages_over) = /.*O V E R.*(\d*).*S T E P S/;
> >>>>>>> }
> >>>>>>>
> >>>>>>> $rms = 1 if (/R M S/);
> >>>>>>>
> >>>>>>> if (/NSTEP/) {
> >>>>>>> ($time, $temp, $pres) =
> >>>>>>> /NSTEP =.*TIME.* =(.*\d*\.\d*).*TEMP.*
> =(.*\d*\.\d*).*PRESS =
> >>>>>>> (.*\d*\.\d*)/;
> >>>>>>> if ( $debug ) {
> >>>>>>> print $_;
> >>>>>>> print "time is $time, temp is $temp, pres is $pres\n";
> >>>>>>> }
> >>>>>>> $_ = <INPUT>;
> >>>>>>>
> >>>>>>> if (/Etot/) {
> >>>>>>> ($etot, $ektot, $eptot) =
> >>>>>>>
> >>>>>>> /Etot.*=(.*\d*\.\d*).*EKtot.*=(.*\d*\.\d*).*EPtot.*=(.*\d*\.\d*)/;
> >>>>>>> if ( $debug ) {
> >>>>>>> print $_;
> >>>>>>> print "Etot is $etot, ektot is $ektot, eptot is
> $eptot\n";
> >>>>>>> }
> >>>>>>> $_ = <INPUT>;
> >>>>>>> }
> >>>>>>> if (/BOND.*ANGLE.*DIHED/) {
> >>>>>>> ($bond, $angle, $dihedral) =
> >>>>>>>
> >>>>>>> /BOND.*=(.*\d*\.\d*).*ANGLE.*=(.*\d*\.\d*).*DIHED.*=(.*\d*\.\d*)/;
> >>>>>>> if ( $debug ) {
> >>>>>>> print $_;
> >>>>>>> print "bond is $bond, angle is $angle, dihedral is
> >>>>>>> $dihedral\n";
> >>>>>>> }
> >>>>>>> $_ = <INPUT>;
> >>>>>>> }
> >>>>>>> if (/1-4 NB/) {
> >>>>>>> ($nb14, $eel14, $nb) =
> >>>>>>> /1-4 NB.*=(.*\d*\.\d*).*1-4
> >>>>>>> EEL.*=(.*\d*\.\d*).*VDWAALS.*=(.*\d*\.\d*)/;
> >>>>>>> if ( $debug ) {
> >>>>>>> print $_;
> >>>>>>> print "nb14 is $nb14, eel14 is $eel14, vdwaals is
> $nb\n";
> >>>>>>> }
> >>>>>>> $_ = <INPUT>;
> >>>>>>> }
> >>>>>>> if (/EELEC/) {
> >>>>>>> ($eel, $ehbond, $constraint) =
> >>>>>>>
> >>>>>>>
> >>>>>>>
> /EELEC.*=(.*\d*\.\d*).*EHBOND.*=(.*\d*\.\d*).*CONSTRAINT.*=(.*\d*\.\d*)/;
> >>>>>>> if ( $debug ) {
> >>>>>>> print $_;
> >>>>>>> print "eel is $eel, ehbond is $ehbond, constraint is
> >>>>>>> $constraint\n";
> >>>>>>> }
> >>>>>>> $_ = <INPUT>;
> >>>>>>> #
> >>>>>>> # check to see if EAMBER is in the mdout file
> (present
> >>>>>>> when
> >>>>>>> # NTR=1)
> >>>>>>> #
> >>>>>>> if ( /EAMBER/ ) {
> >>>>>>> $_ = <INPUT>;
> >>>>>>> }
> >>>>>>> }
> >>>>>>> if (/EKCMT/) {
> >>>>>>> ($ekcmt, $virial, $volume) =
> >>>>>>>
> >>>>>>>
> /EKCMT.*=(.*\d*\.\d*).*VIRIAL.*=(.*\d*\.\d*).*VOLUME.*=(.*\d*\.\d*)/;
> >>>>>>> if ( $debug ) {
> >>>>>>> print $_;
> >>>>>>> print "Ekcmt is $ekcmt, virial is $virial, volume is
> >>>>>>> $volume\n";
> >>>>>>> }
> >>>>>>> $_ = <INPUT>;
> >>>>>>> }
> >>>>>>> if (/T_SOLUTE/) {
> >>>>>>> ($tsolute, $tsolvent) =
> >>>>>>> /T_SOLUTE =(.*\d*\.\d*).*T_SOLVENT =(.*\d*\.\d*)/;
> >>>>>>> if ( $debug ) {
> >>>>>>> print $_;
> >>>>>>> print "Temp solute is $tsolute, temp solvent is
> >>>>>>> $tsolvent\n";
> >>>>>>> }
> >>>>>>> $_ = <INPUT>;
> >>>>>>> }
> >>>>>>>
> >>>>>>> if (/Density/) {
> >>>>>>> ($density) = /.*Density.*=(.*\d*\.\d*)/;
> >>>>>>> if ( $debug ) {
> >>>>>>> print $_;
> >>>>>>> print "Density is $density\n";
> >>>>>>> }
> >>>>>>> $_ = <INPUT>;
> >>>>>>> }
> >>>>>>>
> >>>>>>> if (/Etot/) {
> >>>>>>> ($etot, $ektot, $eptot) =
> >>>>>>>
> >>>>>>> /Etot.*=(.*\d*\.\d*).*EKtot.*=(.*\d*\.\d*).*EPtot.*=(.*\d*\.\d*)/;
> >>>>>>> if ( $debug ) {
> >>>>>>> print $_;
> >>>>>>> print "Etot is $etot, ektot is $ektot, eptot is
> $eptot\n";
> >>>>>>> }
> >>>>>>> $_ = <INPUT>;
> >>>>>>> }
> >>>>>>> if (/ESCF/) {
> >>>>>>> ($escf) =
> >>>>>>> /.*ESCF.*=(.*\d*\.\d*)/;
> >>>>>>> if ( $debug ) {
> >>>>>>> print $_;
> >>>>>>> print "ESCF is $escf\n";
> >>>>>>> }
> >>>>>>> $_ = <INPUT>;
> >>>>>>> }
> >>>>>>>
> >>>>>>> # update arrays
> >>>>>>>
> >>>>>>> if ( $averages == 1 ) {
> >>>>>>> $AVG_TIME{$time} = $time;
> >>>>>>> $AVG_TEMP{$time} = $temp;
> >>>>>>> $AVG_PRES{$time} = $pres;
> >>>>>>> $AVG_ETOT{$time} = $etot;
> >>>>>>> $AVG_EKTOT{$time} = $ektot;
> >>>>>>> $AVG_EPTOT{$time} = $eptot;
> >>>>>>> $AVG_BOND{$time} = $bond;
> >>>>>>> $AVG_ANGLE{$time} = $angle;
> >>>>>>> $AVG_DIHEDRAL{$time} = $dihedral;
> >>>>>>> $AVG_NB14{$time} = $nb14;
> >>>>>>> $AVG_EEL14{$time} = $eel14;
> >>>>>>> $AVG_NB{$time} = $nb;
> >>>>>>> $AVG_EEL{$time} = $eel;
> >>>>>>> $AVG_EHBOND{$time} = $ehbond;
> >>>>>>> $AVG_CONSTRAINT{$time} = $constraint;
> >>>>>>> $AVG_EKCMT{$time} = $ekcmt;
> >>>>>>> $AVG_VIRIAL{$time} = $virial;
> >>>>>>> $AVG_VOLUME{$time} = $volume;
> >>>>>>> $AVG_TSOLUTE{$time} = $tsolute;
> >>>>>>> $AVG_TSOLVENT{$time} = $tsolvent;
> >>>>>>> $AVG_DENSITY{$time} = $density;
> >>>>>>> $AVG_ESCF{$time} = $escf;
> >>>>>>> $averages = 0;
> >>>>>>> } elsif ( $rms == 1 ) {
> >>>>>>> $RMS_TIME{$time} = $time;
> >>>>>>> $RMS_TEMP{$time} = $temp;
> >>>>>>> $RMS_PRES{$time} = $pres;
> >>>>>>> $RMS_ETOT{$time} = $etot;
> >>>>>>> $RMS_EKTOT{$time} = $ektot;
> >>>>>>> $RMS_EPTOT{$time} = $eptot;
> >>>>>>> $RMS_BOND{$time} = $bond;
> >>>>>>> $RMS_ANGLE{$time} = $angle;
> >>>>>>> $RMS_DIHEDRAL{$time} = $dihedral;
> >>>>>>> $RMS_NB14{$time} = $nb14;
> >>>>>>> $RMS_EEL14{$time} = $eel14;
> >>>>>>> $RMS_NB{$time} = $nb;
> >>>>>>> $RMS_EEL{$time} = $eel;
> >>>>>>> $RMS_EHBOND{$time} = $ehbond;
> >>>>>>> $RMS_CONSTRAINT{$time} = $constraint;
> >>>>>>> $RMS_EKCMT{$time} = $ekcmt;
> >>>>>>> $RMS_VIRIAL{$time} = $virial;
> >>>>>>> $RMS_VOLUME{$time} = $volume;
> >>>>>>> $RMS_TSOLUTE{$time} = $tsolute;
> >>>>>>> $RMS_TSOLVENT{$time} = $tsolvent;
> >>>>>>> $RMS_DENSITY{$time} = $density;
> >>>>>>> $RMS_ESCF{$time} = $escf;
> >>>>>>>
> >>>>>>> $rms = 0;
> >>>>>>> } else {
> >>>>>>> $TIME{$time} = $time;
> >>>>>>> $TEMP{$time} = $temp;
> >>>>>>> $PRES{$time} = $pres;
> >>>>>>> $ETOT{$time} = $etot;
> >>>>>>> $EKTOT{$time} = $ektot;
> >>>>>>> $EPTOT{$time} = $eptot;
> >>>>>>> $BOND{$time} = $bond;
> >>>>>>> $ANGLE{$time} = $angle;
> >>>>>>> $DIHEDRAL{$time} = $dihedral;
> >>>>>>> $NB14{$time} = $nb14;
> >>>>>>> $EEL14{$time} = $eel14;
> >>>>>>> $NB{$time} = $nb;
> >>>>>>> $EEL{$time} = $eel;
> >>>>>>> $EHBOND{$time} = $ehbond;
> >>>>>>> $CONSTRAINT{$time} = $constraint;
> >>>>>>> $EKCMT{$time} = $ekcmt;
> >>>>>>> $VIRIAL{$time} = $virial;
> >>>>>>> $VOLUME{$time} = $volume;
> >>>>>>> $TSOLUTE{$time} = $tsolute;
> >>>>>>> $TSOLVENT{$time} = $tsolvent;
> >>>>>>> $DENSITY{$time} = $density;
> >>>>>>> $ESCF{$time} = $escf;
> >>>>>>> }
> >>>>>>>
> >>>>>>> }
> >>>>>>> }
> >>>>>>> }
> >>>>>>>
> >>>>>>> But after running this perl script on the output files I got graph
> >>>>>>> displaying half values of output files as attached here. Can
> anybody
> >>>>>> guide
> >>>>>>> me whats wrong with the perl graph or my input file as my output
> files
> >>>>>> are
> >>>>>>> printing all values for energy but perl script plot half values???
> >>>>>>>
> >>>>>>>
> >>>>>>>
> >>>>>>> Sent from Mail for Windows 10
> >>>>>>>
> >>>>>>>
> >>>>>>> _______________________________________________
> >>>>>>> AMBER mailing list
> >>>>>>> AMBER.ambermd.org
> <javascript:_e(%7B%7D,'cvml','AMBER.ambermd.org');>
> >>>>>>> http://lists.ambermd.org/mailman/listinfo/amber
> >>>>>>>
> >>>>>>>
> >>>>>> _______________________________________________
> >>>>>> AMBER mailing list
> >>>>>> AMBER.ambermd.org
> <javascript:_e(%7B%7D,'cvml','AMBER.ambermd.org');>
> >>>>>> http://lists.ambermd.org/mailman/listinfo/amber
> >>>>>> _______________________________________________
> >>>>>> AMBER mailing list
> >>>>>> AMBER.ambermd.org
> <javascript:_e(%7B%7D,'cvml','AMBER.ambermd.org');>
> >>>>>> http://lists.ambermd.org/mailman/listinfo/amber
> >>>>>>
> >>>>> _______________________________________________
> >>>>> AMBER mailing list
> >>>>> AMBER.ambermd.org
> <javascript:_e(%7B%7D,'cvml','AMBER.ambermd.org');>
> >>>>> http://lists.ambermd.org/mailman/listinfo/amber
> >>>>> _______________________________________________
> >>>>> AMBER mailing list
> >>>>> AMBER.ambermd.org
> <javascript:_e(%7B%7D,'cvml','AMBER.ambermd.org');>
> >>>>> http://lists.ambermd.org/mailman/listinfo/amber
> >>>> _______________________________________________
> >>>> AMBER mailing list
> >>>> AMBER.ambermd.org <javascript:_e(%7B%7D,'cvml','AMBER.ambermd.org');>
> >>>> http://lists.ambermd.org/mailman/listinfo/amber
> >>>> _______________________________________________
> >>>> AMBER mailing list
> >>>> AMBER.ambermd.org <javascript:_e(%7B%7D,'cvml','AMBER.ambermd.org');>
> >>>> http://lists.ambermd.org/mailman/listinfo/amber
> >>>
> >>> --
> >>> -------------------------
> >>> Daniel R. Roe, PhD
> >>> Department of Medicinal Chemistry
> >>> University of Utah
> >>> 30 South 2000 East, Room 307
> >>> Salt Lake City, UT 84112-5820
> >>> http://home.chpc.utah.edu/~cheatham/
> >>> (801) 587-9652
> >>> (801) 585-6208 (Fax)
> >>>
> >>> _______________________________________________
> >>> AMBER mailing list
> >>> AMBER.ambermd.org <javascript:_e(%7B%7D,'cvml','AMBER.ambermd.org');>
> >>> http://lists.ambermd.org/mailman/listinfo/amber
> >>
> >> --
> >> -------------------------
> >> Daniel R. Roe, PhD
> >> Department of Medicinal Chemistry
> >> University of Utah
> >> 30 South 2000 East, Room 307
> >> Salt Lake City, UT 84112-5820
> >> http://home.chpc.utah.edu/~cheatham/
> >> (801) 587-9652
> >> (801) 585-6208 (Fax)
> >>
> >> _______________________________________________
> >> AMBER mailing list
> >> AMBER.ambermd.org <javascript:_e(%7B%7D,'cvml','AMBER.ambermd.org');>
> >> http://lists.ambermd.org/mailman/listinfo/amber
> >> _______________________________________________
> >> AMBER mailing list
> >> AMBER.ambermd.org <javascript:_e(%7B%7D,'cvml','AMBER.ambermd.org');>
> >> http://lists.ambermd.org/mailman/listinfo/amber
> >
> > _______________________________________________
> > AMBER mailing list
> > AMBER.ambermd.org <javascript:_e(%7B%7D,'cvml','AMBER.ambermd.org');>
> > http://lists.ambermd.org/mailman/listinfo/amber
>
>
> _______________________________________________
> AMBER mailing list
> AMBER.ambermd.org <javascript:_e(%7B%7D,'cvml','AMBER.ambermd.org');>
> http://lists.ambermd.org/mailman/listinfo/amber
> _______________________________________________
> AMBER mailing list
> AMBER.ambermd.org <javascript:_e(%7B%7D,'cvml','AMBER.ambermd.org');>
> http://lists.ambermd.org/mailman/listinfo/amber
>
-- ------------------------- Daniel R. Roe, PhD Department of Medicinal Chemistry University of Utah 30 South 2000 East, Room 307 Salt Lake City, UT 84112-5820 http://home.chpc.utah.edu/~cheatham/ (801) 587-9652 (801) 585-6208 (Fax) _______________________________________________ AMBER mailing list AMBER.ambermd.org http://lists.ambermd.org/mailman/listinfo/amber
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: energy_tot.png)
(image/png attachment: energy_incomplete.png)
