Date: Tue, 29 Mar 2016 20:05:05 +0800
Hello all,
An MD simulation in Amber14 looks weird in VMD 1.9.2 but looks perfectly
fine in Chimera.
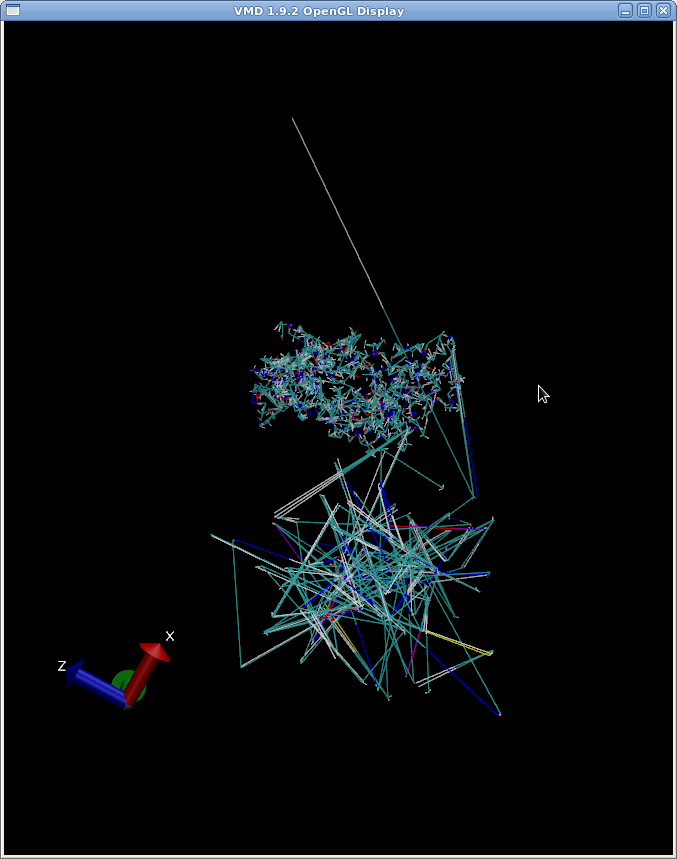
There seems to be a "tail" and some protruding parts that appear to trace a
path below (and sometimes in other places elsewhere) the protein being
simulated (this is part of the equilibriation stage). Please see attached
photo.
Any ideas/ advice? Thanks!
--
Cheers,
Enrique
<http://www.rbc.org/odb/odb.shtml>
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: trajectory_artefact_amber_viewed_in_VMD1-9-2.png)
