Date: Wed, 27 Jan 2016 11:05:38 +0000
Dear all,
I am facing very bizarre problem while loading my protein in prmtop and inpcrd files in VMD. Two of the bonds are extraordinarily elongated when we open the files in Amber trajectory with periodic box whereas the same files open in amber restart format (file type) is ok. Secondly, the problem is occuring only in prmtop and inpcrd visulaization while the pdb generated and saved from xleap module after the generation of prmtop and inpcrd was also normal. Attach are the snapshots of the above mentioned file for your visulaization. Please kindly give suggestions accordingly.
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
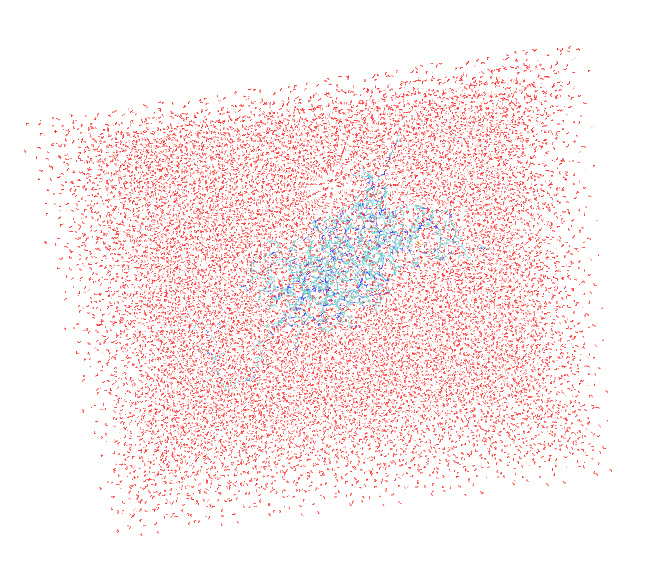
(image/png attachment: Box_rstformat.png)
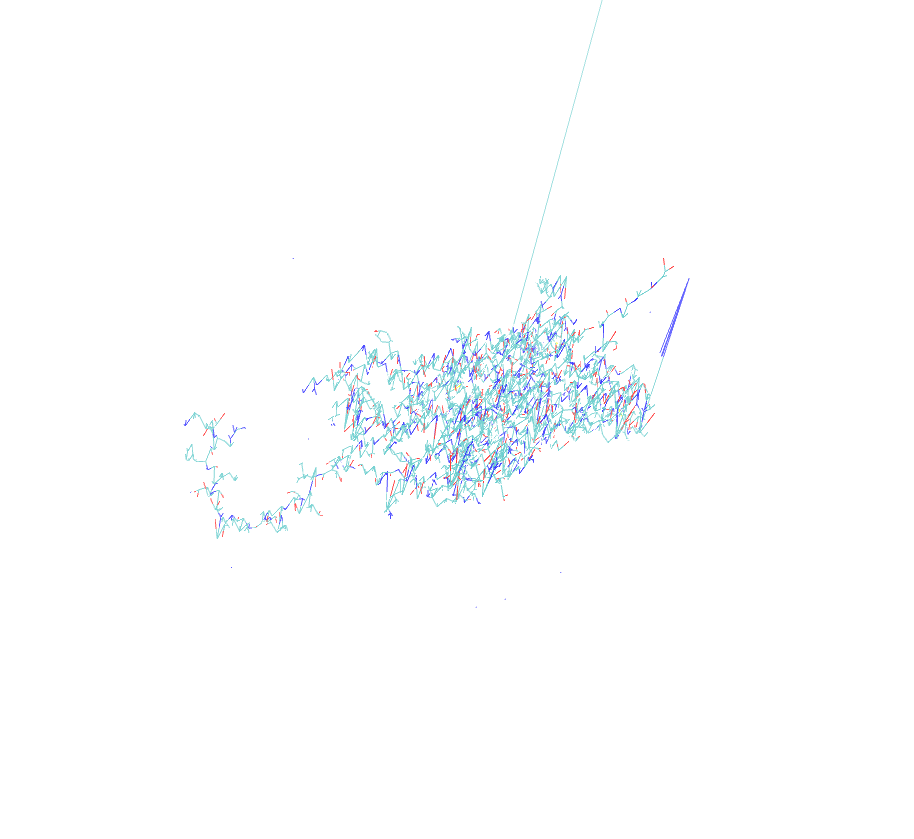
(image/png attachment: protein_elongated.png)
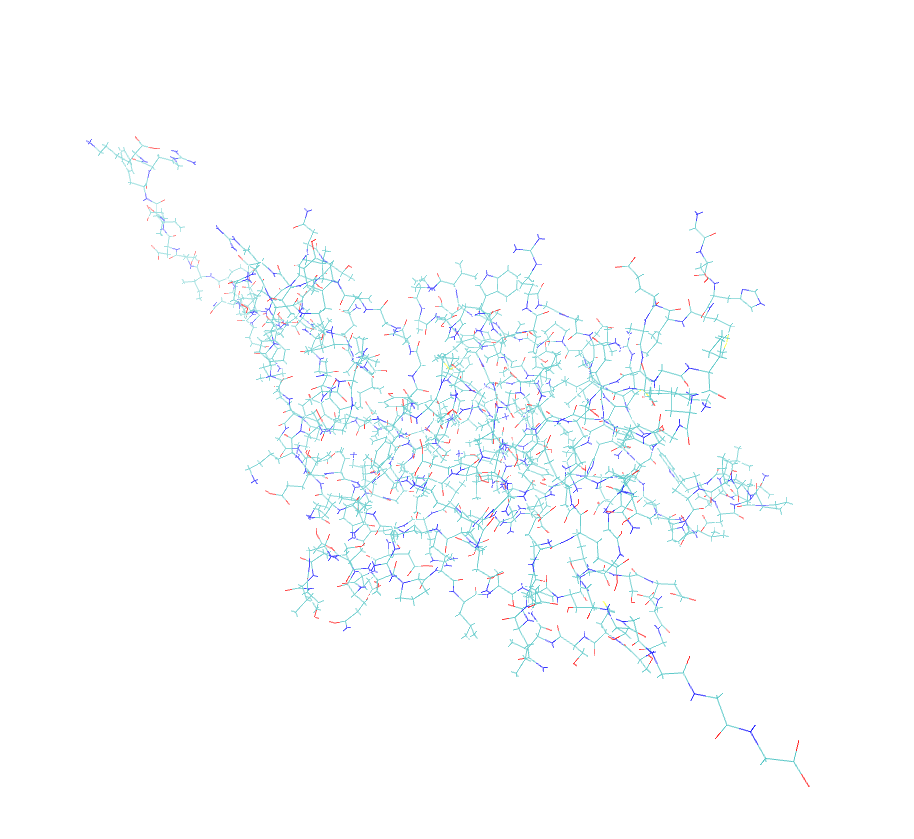
(image/png attachment: protein_pdb.png)
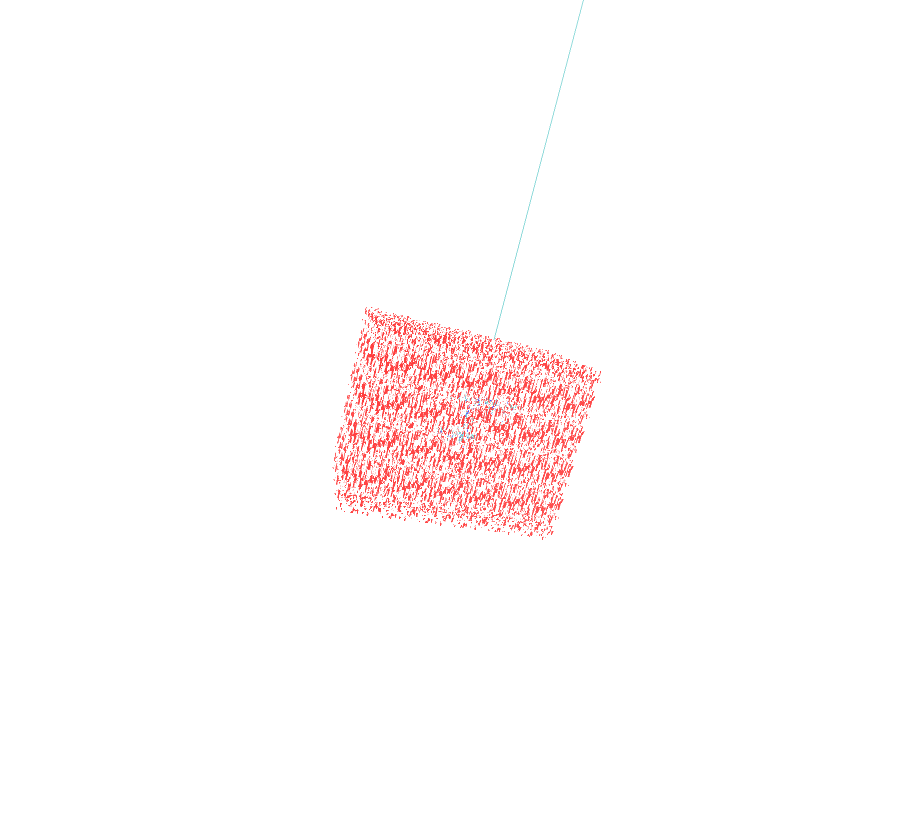
(image/png attachment: waterbox_elongatedprotein.png)
