Date: Mon, 27 Jul 2015 13:32:23 +0300
Greetings,
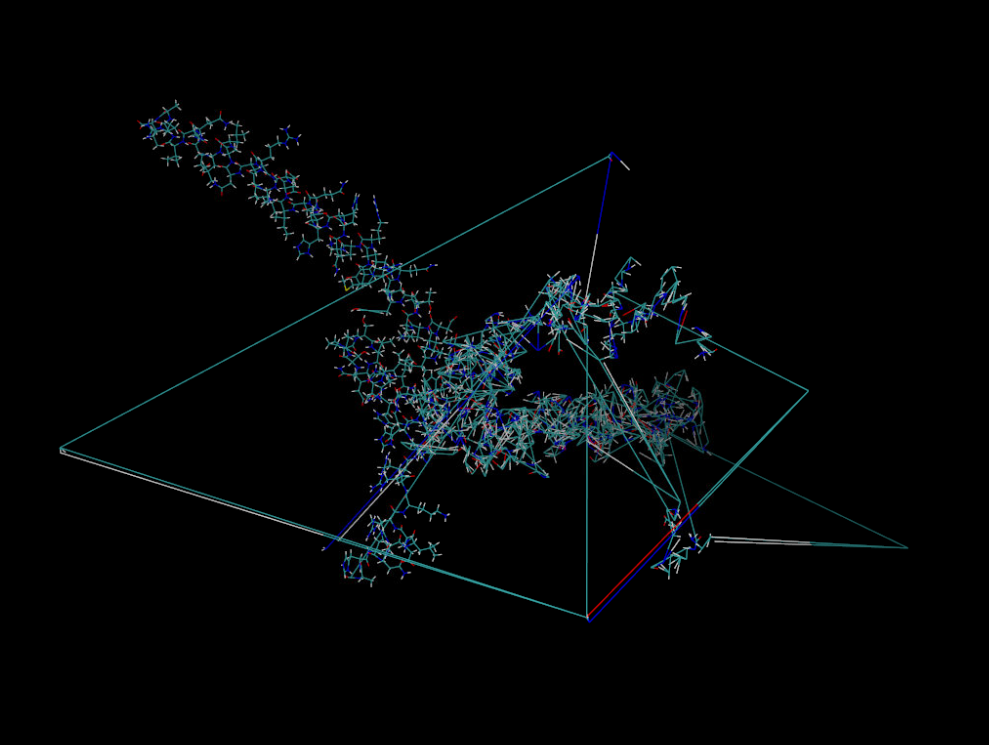
I have a long netcdf trajectory with several corrupted frames like the
attached one. The command lines I used in cpptraj to process the
trajectories are the following:
strip !:1-162
center :1-162 mass origin
image origin center familiar
trajout protein.box.noimage.nc netcdf nobox
trajout first_frm_protein.pdb pdb onlyframes 1
Residues 1-162 correspond to the protein. I have two questions in this
regard:
1) Is there any simple way to detect corrupted frames with cpptraj?
2) Can I specify a list of frames that I want to be discarded by cpptraj
when it reads the processed trajectory?
thanks,
Thomas
--
======================================================================
Thomas Evangelidis
Research Specialist
CEITEC - Central European Institute of Technology
Masaryk University
Kamenice 5/A35/1S081,
62500 Brno, Czech Republic
email: tevang.pharm.uoa.gr
tevang3.gmail.com
website: https://sites.google.com/site/thomasevangelidishomepage/
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: corrupted.png)
