Date: Thu, 19 Mar 2015 11:40:59 +0800
Dear Amber users,
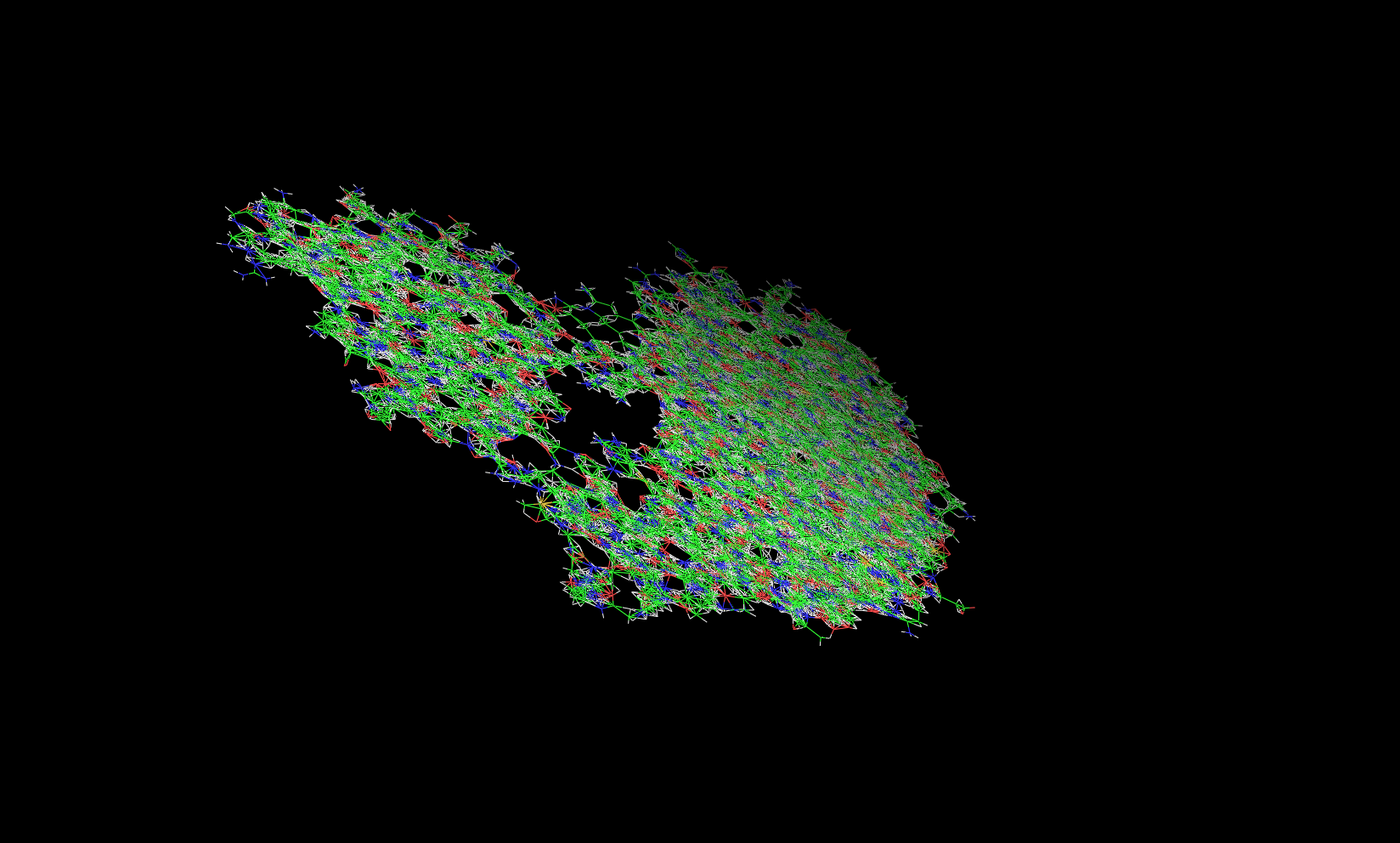
I used cpptraj to convert a trajectory frame into pdb format, but I got
very "weird" pdb file (Sorry, I cannot attach it here as it is more than
5Mb and the post was bounced back). When I visualized it, it gave me a
"flat" structure with clashes (image attached).
Could you please help comment if I missed something? Thanks.
> parm gagIT.prmtop
Reading 'gagIT.prmtop' as Amber Topology
> trajin gagIT_md950ns.mdcrd
Reading 'gagIT_md950ns.mdcrd' as Amber Trajectory
> trajout gagIT_md950ns.pdb pdb parm gagIT.prmtop onlyframes 2500
Writing 'gagIT_md950ns.pdb' as PDB
Saving frames 2500
> run
---------- RUN BEGIN -------------------------------------------------
PARAMETER FILES:
0: 'gagIT.prmtop', 59573 atoms, 17761 res, box: Trunc. Oct., 17262 mol,
17248 solvent, 2500 frames
INPUT TRAJECTORIES:
0: 'gagIT_md950ns.mdcrd' is an AMBER trajectory, Parm gagIT.prmtop (Trunc.
Oct. box) (reading 2500 of 2500)
Coordinate processing will occur on 2500 frames.
OUTPUT TRAJECTORIES:
'gagIT_md950ns.pdb' is a PDB file, Parm gagIT.prmtop: Writing frames 2500
TIME: Run Initialization took 0.0001 seconds.
BEGIN TRAJECTORY PROCESSING:
----- gagIT_md950ns.mdcrd (1-2500, 1) -----
0% 10% 20% 30% 40% 50% 60% 70% 80% 90% 100% Complete.
Read 2500 frames and processed 2500 frames.
TIME: Trajectory processing: 49.1169 s
TIME: Avg. throughput= 50.8989 frames / second.
ACTION OUTPUT:
---------- RUN END ---------------------------------------------------
_______________________________________________
AMBER mailing list
AMBER.ambermd.org
http://lists.ambermd.org/mailman/listinfo/amber
(image/png attachment: weirdPDB.png)
